Abstract
The CRISPR/Cas9 system provides a novel and promising tool for editing the HIV-1 proviral genome. We designed RNA-guided CRISPR/Cas9 targeting the HIV-1 regulatory genes tat and rev with guide RNAs (gRNA) selected from each gene based on CRISPR specificity and sequence conservation across six major HIV-1 subtypes. Each gRNA was cloned into lentiCRISPRv2 before co-transfection to create a lentiviral vector and transduction into target cells. CRISPR/Cas9 transduction into 293 T and HeLa cells stably expressing Tat and Rev proteins successfully abolished the expression of each protein relative to that in non-transduced and gRNA-absent vector-transduced cells. Tat functional assays showed significantly reduced HIV-1 promoter-driven luciferase expression after tat-CRISPR transduction, while Rev functional assays revealed abolished gp120 expression after rev-CRISPR transduction. The target gene was mutated at the Cas9 cleavage site with high frequency and various indel mutations. Conversely, no mutations were detected at off-target sites and Cas9 expression had no effect on cell viability. CRISPR/Cas9 was further tested in persistently and latently HIV-1-infected T-cell lines, in which p24 levels were significantly suppressed even after cytokine reactivation, and multiplexing all six gRNAs further increased efficiency. Thus, the CRISPR/Cas9 system targeting HIV-1 regulatory genes may serve as a favorable means to achieve functional cures.
Similar content being viewed by others
Introduction
Human immunodeficiency virus-1 (HIV-1) infection continues to be a major health issue with more than 35 million individuals being infected worldwide1. Combination antiretroviral therapy (ART) has been effective at suppressing viremia and restoring primary host CD4+ T cells as well as improving clinical outcomes, thereby transforming a deadly infection into a manageable chronic disease2. However, low-level virion production continues to exist due to viral persistence inside cellular (e.g. resting CD4+ T cells, macrophages, and follicular dendritic cells) and anatomical reservoirs (e.g. the central nervous system)3. Among these, resting CD4+ T cells have been the most extensively studied and are considered to be the major obstacle to HIV-1 eradication; a small fraction (0.2–16.4/106) of resting CD4+ T cells harbors latent HIV-1 even under ART suppression4,5. Current antiviral compounds are incapable of targeting the integrated proviral genome inside these cellular reservoirs and rapid viral rebound ensues after ART cessation6,7,8. Life-long treatment is mandatory, yet long-term medication has also been complicated by issues such as the chronic side effects of ART, adherence difficulties, the emergence of escape mutants, and morbidities in aging populations due to the prolonged life expectancy of people living with HIV-19,10,11,12. A single-time intervention to clear viral genomic persistence represents a sensible and promising approach to achieve a HIV-1 functional cure. In latently infected CD4+ T cells, the proviral genome may shift into a dormant state in which inert transcription causes almost no production of viral proteins, and may later be reactivated during T-cell activation to assemble replication-competent viruses13. One strategy is to deliberately induce this reactivation, termed the “shock and kill” approach, using latency-reversing agents such as inhibitors of histone deacetylase (HDACi), and this method has shown the potent induction of T-cell activation followed by viral mRNA increases14; however, ex vivo assays suggested that the outgrowth of HIV-1 from latent reservoirs is insufficient and cytotoxic T-cell responses are not sufficiently strong to eliminate reactivated infected cells, resulting in a minimal impact on the overall reservoir size15,16. These findings indicate that strategies for a HIV-1 cure need to involve the direct disruption of the proviral genome from the cellular reservoir, which may be achieved with site-specific genome editing.
Over the last two decades, advances have been achieved in genome editing technology through the innovation of site-directed engineered nucleases, such as zinc finger nuclease (ZFN) and transcription activator-like effector nuclease (TALEN), which uses the DNA-protein recognition principle to direct FokI nuclease towards essentially any sequence within the genome and digest it17,18. However, difficulties associated with design, synthesis, and protein validation for a specific gene locus of interest have restricted the feasibility of these methods19. A key breakthrough was made when a bacterial immune system-related RNA molecule, called the clustered regularly interspaced short palindromic repeats (CRISPR), was found to be able to guide CRISPR-associated 9 (Cas9) nuclease towards DNA sequences matching those of the guide RNA (gRNA). This gRNA is easily programmable and the simple transduction of the designed gRNA with a Cas9 expression cassette may introduce double-strand breaks (DSB) inside DNA in a highly specific and efficient manner20. CRISPR also has the advantage over ZFN and TALEN of being a smaller size and, thus, is easier to package into lentiviral constructs, has a lower risk of off-target cleavage, is easier to create, less costly, and has demonstrated higher efficiency19,21. The CRISPR/Cas9 system has achieved successful outcomes in many mammalian culture cells, including human T-cell lines22 and pluripotent cell lines, and has been tried-and-tested in a broad range of in vitro and in vivo studies on human genetic and infectious diseases23,24, including HIV-125.
The successful late transcription of HIV-1 following viral activation is highly dependent on the early expression of the regulatory proteins Tat and Rev. The elongation of nascent viral mRNA from the integrated provirus is initiated by Tat, while the nuclear export of unspliced transcripts is regulated by Rev26,27. In HIV-1-infected activated T cells, the combination of Tat and Rev provide a very high level of viral gene expression, while the same proteins in resting T cells are important for maintaining the provirus in a latent state28. tat and rev are considered to be some of the most functionally conserved genes of HIV-1, with some genomic domains inside sharing the same homology across wide HIV-1 subtypes and even to HIV-2 and simian immunodeficiency virus (SIV)28,29. Many RNA-based30,31,32,33,34,35 and protein-based33,36,37,38 anti-HIV-1 moieties targeting these proteins or their exons have been successfully shown to reduce viral replication in T cells to a varying degree with methods including, but not being limited to Tat/Rev short hairpin RNA (shRNA), antisense RNA, a trans-activation response/Rev response element (TAR/RRE) decoy, mutant molecules, and tat/rev-targeting ribozymes. CRISPR/Cas9 provides a promising new method to target the regulatory genes of HIV-1. In the present study, we constructed a lentiviral vector-based CRISPR/Cas9 system harboring gRNAs with the ability to recognize specific DNA sequences inside the coding sequences of Tat and Rev. We demonstrated that the CRISPR/Cas9 system abolished Tat and Rev protein expression and their respective regulatory functions in stable Tat- and Rev-expressing 293 T cells. Target-site sequencing confirmed that the Cas9-associated mutation occurred inside the tat and rev exons, while no off-target mutations were detected in sequences similar to the designed gRNAs inside the human genome. We ultimately found that CRISPR transduction successfully diminished viral capsid production in persistently and latently infected CD4+ T-cell lines. These results support the potential use of CRISPR to specifically target HIV-1 regulatory genes and suppress viral replication.
Results
CRISPR/Cas9 abolished the expression and function of Tat and Rev proteins
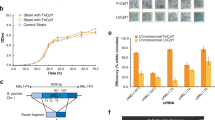
We designed six gRNAs with three constructs targeting each tat and rev gene (Fig. 1A). All gRNAs contained a 20-bp sequence from targeted genes followed by the 3-bp CRISPR recognition site, called the protospacer adjacent motif (PAM, NGG). Three tat-targeting gRNAs were taken from the first exon of tat with tatA targeting the N-terminal acidic domain, tatB targeting the short core domain hitting the highly conserved RKGLGI motif, and tatC targeting the end of the acidic domain to the start of cysteine residues. Three rev-targeting gRNAs were taken from the second exon of rev with revA targeting the arginine-rich motif in the nuclear localization signal and the RNA-binding domain, revB targeting the second multimerization domain, which is crucial for forming the alpha-helical secondary structure of the Rev protein, and revC targeting the leucine-rich nuclear export signal effector domain. CRISPR specificity scores were counted based on on-target activity minus the weighted-sum of off-target probability as calculated by software in http://crispr.mit.edu. Selected sequences have relatively high hit scores in this algorithm. The designed 23-bp gRNAs were aligned to 97 HIV-1 strains from subtypes A, B, C, D, CRF01AE, and CRF02AG as curated from the Los Alamos National Laboratory (LANL) database and showed relatively high sequence homology at each site (Fig. 1B).
CRISPR gRNAs targeting HIV-1 regulatory genes designed and tested in this study. (A) Schematic representation of the HIV-1 proviral genome with the approximate location of each gRNA. Three target sequences were selected from each tat and rev region: tatA, tatB, tatC, revA, revB, and revC. The specific domains targeted in each gene are shown. (B) Sequences of gRNAs with the PAM region highlighted in red and the Cas9 cleavage site 3-nt upstream of PAM denoted by a red line, and respective coordinates in HXB2. CRISPR specificity scores were calculated by algorithms available in http://crispr.mit.edu and the conservation of each sequence was performed across 97 HIV-1 strains of six subtypes (A, B, C, D, 01AE, and 02AG) as curated from the Los Alamos National Lab (LANL) HIV Sequence Compendium. Conservation percentages and nucleotide variabilities as measured by Shannon entropy are shown.
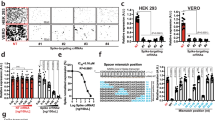
In order to analyze the effects of the designed CRISPR/Cas9 construct to HIV-1 regulatory protein expression, we first developed stable, FLAG-bound Tat- and Rev-expressing HEK 293 T cell lines by infecting cells with a lentiviral vector bearing FLAG-tagged HIV-1 NL4-3 tat and rev, mimicking actual HIV-1 infection. These transformant cells constantly produced FLAG-bound Tat and Rev proteins at high amounts, as confirmed by an anti-FLAG immunoblot assay. Vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped lentiviral vectors expressing S. pyogenes Cas9 nuclease bearing the designed gRNAs were then transduced into 293T-Tat and 293T-Rev cells, either once or twice with a multiplicity of infection (MOI) of 10. MOI of 10 was selected because it was previously shown to be the most optimal for expressing transgenes into target T cells with minimal cytotoxicity39. In addition, the designed gRNAs were not able to target the HIV-1-based lentiviral vectors used for transduction. The lentiviral backbone lentiCRISPRv2 (LC) harbored long terminal repeat (LTR) and RRE genes, but did not regulate the expression of U6 transcription-based gRNA or Cas9. Medium was maintained for five days for each transduction and the cell lysate was then assayed using an immunoblot analysis. Western blot results showed that tat- and rev-CRISPR abolished Tat and Rev expression relative to that in non-transduced cells (WT). A second round of transduction further reduced protein expression (Fig. 2A). A marked reduction in FLAG expression was not observed in cells transduced either once or twice with empty LC, which bore the Cas9 construct without gRNA. On the other hand, the Cas9 protein was expressed in all transduced cells, including empty LC-transduced cells, although expression levels varied to some extent. The same treatment of stable FLAG-Tat- and Rev-expressing HeLa cells provided similar results (see Supplementary Fig. S1). Third-round transduction did not result in further marked reductions in Tat-FLAG protein expression or HIV-1 LTR-driven luciferase transcriptional activity (data not shown); however, two-round transduction resulted in a low level of protein expression.
CRISPR/Cas9 abolished Tat and Rev protein expression and function in stable Tat- and Rev-expressing transformant 293 T cells. (A) Western blot analysis for FLAG-bound Tat and Rev protein expression. Transformant 293 T cells were transduced with a CRISPR/Cas9-bearing lentiviral vector for 3 days before being lysed and immunostained with an anti-FLAG antibody. The transduction of lentiCRISPR without gRNA (empty LC) and with tat/rev-targeting gRNAs was performed once or twice, and protein expression was compared with non-transduced cells (WT). The Cas9 protein was detected by an anti-Cas9 antibody and expression was shown for each cell group. Beta-actin expression was used as the loading control. Assays were performed at least twice for the expression of each protein and the most representative image was selected. Original unedited blots are shown in the supplementary dataset and images were not joined from different parts of the gel. (B) A Tat functional analysis of 293T-Tat cells. WT and CRISPR-transduced cells were transfected with pLTR-Luc for 3 days and cell lysates were obtained to measure luciferase activity. The transfections of pRc/RSV and the Tat expression vector pRSV-Tat into original 293 T cells were used as negative and positive controls, respectively. A one-way ANOVA applied for five groups (WT, empty LC, tatA, tatB, and tatC) gave a p value < 0.001. Pairwise comparisons between WT and CRISPR-transduced cells are shown. Experiments were performed three times and average values ±SE are shown. (C) A Rev functional analysis of 293T-Rev cells. WT and CRISPR-transduced cells were transfected with the gp140-expressing plasmid for 3 days and the cell lysate was immunostained with an anti-gp120 antibody. The transfections of pcDNA3.1/Zeo(+) and the Rev expression plasmid pCA-Rev into original 293 T cells were used as negative and positive controls, respectively. Assays were performed twice and the most representative image was selected. Original unedited blots are shown in the supplementary dataset and images were not joined from different parts of the gel.
The high-level activation of HIV-1 RNA polymerase depends on the binding of Tat-associated kinase and the TAR element inside HIV-1 3′LTR. We used this principle to perform a functional assay on the Tat protein. Previously established 293T-Tat transformant cells, either non-transduced or transduced with empty LC or tat-CRISPR, were transfected with pLTR-Luc for 3 days before being lysed to measure luciferase activity. The plasmid pLTR-Luc contained HIV-1 3′LTR, which drives luciferase expression upon transactivation by the Tat protein onto TAR. The luciferase assay showed that tat-CRISPR significantly suppressed HIV-1 LTR-driven expression in 293T-Tat cells by 61%, 76%, and 77% for tatA, tatB, and tatC (P = 0.024, P = 0.01, and P = 0.012), respectively (Fig. 2B). A similar assay using HeLa-Tat cells showed even stronger suppression: 97%, 97%, and 94% (P = 0.007, P = 0.007, and P = 0.008, see Supplementary Fig. S1), respectively, presumably due to lower transduction efficiency during the establishment of transformant HeLa cells and, thus, weaker initial Tat expression. Conversely, a Rev functional assay was performed by comparing gp120 envelope expression because the nuclear export of incompletely spliced env mRNA depends on the binding of Rev and the RRE RNA stem loop inside env. Control and CRISPR-transduced 293T-Rev cells were transfected with the gp140-expressing plasmid for 3 days before being lysed and immunostained with an anti-gp120 antibody. Western blotting showed the abolished expression of gp120 in 293 T cells treated with rev-CRISPR constructs relative to that in the control and cells transduced with empty LC (Fig. 2C), which was, thus, attributed to failed env mRNA export due to Rev protein dysfunction. Overall, the results of functional assays on the Tat and Rev proteins appeared to be in agreement with FLAG-Tat and Rev-FLAG expression levels.
High frequencies of various types of mutations were found precisely at the site of Cas9 DSB
In order to directly confirm the nuclease activity of CRISPR/Cas9 inside the HIV-1 proviral genome, we compared the on-target base sequences of CRISPR-transduced cells with a reference. The target regions of all six gRNA from tat- and rev-CRISPR were amplified from the extracted DNA of CRISPR-transduced 293 T cells to create plasmids, with at least 20 clones for each gRNA construct, and amplicons were subjected to Sanger sequencing (Fig. 3A). Alignment with the NL4-3 wild-type sequence showed that 106/110 clones (96.4%) for tat and 58/60 (96.7%) clones for rev had mutations. A sequence analysis showed 86 and 16 types of unique patterns of mutations in tat and rev-CRISPR amplicons, respectively, with 77/106 (72.6%) for tat and 56/58 (96.6%) for rev being frameshift mutations. Conversely, no mutations were found in the same number of plasmid clones extracted from wild-type cells and empty LC-transduced cellular DNA. Mutations in CRISPR-transduced cells were mostly deletions (63% and 75% for tat and rev, respectively) followed by insertions (13%, 19%), indel combinations (19%, 6%), and substitutions (5%, 0%), as charted in Fig. 3B. These mutation types are typically observed in the non-homologous end joining (NHEJ)-dependent repair of DNA DSBs, often expected as a result of genome editing without any extrachromosomal template, and the high frequency of deletions was consistent with the patterns observed in CRISPR/Cas9-induced mutations40. The indel sites of all mutated samples were located 3-nt upstream of the PAM recognition site (red vertical line in Fig. 3A), which was the Cas9 nuclease cleavage site. Collectively, these results strongly indicated that NHEJ repair-induced mutations found inside CRISPR-transduced cells were caused by CRISPR/Cas9 enzymatic activity.
Sequencing analysis of the on-target site for each CRISPR gRNA. (A) On-target sites were amplified from the extracted DNA of CRISPR-transduced 293 T cells in order to create at least 20 plasmid clones for each gRNA construct, and amplicons were Sanger sequenced. Wild-type NL4-3 sequences are shown topmost with the PAM site (NGG) indicated in blue. Mutations are indicated in red and frameshift mutations are denoted with a blue asterisk (*). Cas9 cleavage sites are indicated in red vertical lines. Percentage numbers at the left side of each sequence denote the frequency of the corresponding mutation pattern found within all sequenced samples for each gRNA. (B) Mutation frequency and distribution of mutation types for each gRNA, cumulative for tat-CRISPR and rev-CRISPR, and overall CRISPR-transduced samples.
CRISPR/Cas9 transduction showed no changes in cell viability and no detectable off-target mutations on the human genome
The recognition mechanism of CRISPR/Cas9 is known to tolerate single and double sequence mismatches between gRNA and target DNA, depending on the position along the gRNA-DNA interface, but particularly in nucleotides distal to the PAM region, and promotes undesired off-target mutations that may introduce genotoxicity and impair cell viability41. A cell viability assay was conducted in order to examine the effects of our CRISPR/Cas9 construct on T cells. Wild-type, empty LC-transduced, and CRISPR-transduced L-2 cells were plated at the same density and subsequent cell growth was monitored for up to 6 weeks; no marked differences were found in cell counts using trypan blue exclusion (Fig. 4, left panel). We also investigated the probability of off-target digestion for each gRNA inside the cellular genome by taking into account the total number of nucleotide mismatches, mean pairwise distance between mismatches, and base distance to the PAM recognition site as calculated by the algorithm in http://crispr.mit.edu. Two off-target sites per gRNA with the highest predicted probability of mutations (Supplementary Table 1) were amplified from cellular DNA extracted from CRISPR-transduced cells. Amplicons were then hybridized to form homo- and heteroduplexes and screened using a Surveyor mutation detection assay, in which the presence of any mutation will show as Surveyor nuclease cleavage products with higher mobility inside an agarose gel than the original amplicon. We herein demonstrated that the gel electrophoresis of off-target amplicons for each gRNA showed no apparent cleavage product, while tat and rev on-target control amplicons both showed the presence of mutations (Fig. 4, right panel).
CRISPR/Cas9 transduction showed no changes in cell viability or detectable off-target activity in the human genome. (Left) Wild-type, empty lentiCRISPR-transduced, and CRISPR-transduced (blue, green, and yellow lines, respectively) L-2 CD4+ T cells were plated at the same density, cell growth was monitored for up to 6 weeks, and viable cells were counted using trypan blue exclusion. Data for each gRNA construct are shown. Experiments were performed three times with average values ±SE as shown. (Right) Surveyor mutation detection assays of two off-target sites in the human genome with the highest predicted probability per gRNA construct and corresponding on-target sites as the positive control. A detailed list of off-target sites is shown in Supplementary Table 1. Assays were performed twice for each gRNA and the most representative image was selected. Images were not joined from different parts of the gel.
CRISPR/Cas9 suppressed HIV-1 replication in persistently and latently infected human CD4+ T-cell lines
L-2 cells are a human CD4+ T-cell line harboring the LAV strain of HIV-1 and persistently release a defective p24 capsid protein into the supernatant42. gRNA/CRISPR constructs were transduced into these cells for 5 days with MOI 10, the culture was changed to fresh medium, and p24 production in the supernatant was measured to compare HIV-1 replication. Since combination therapy is considered to increase inhibition potency, multiple gRNA transductions with a combination cocktail of three tat and rev gRNA-bearing lentiviral vectors (tatABC and revABC) and six gRNAs (tatrevABC) were also performed. Lentivirus concentrations and MOI were maintained in all multiplex gRNA treatment groups, i.e. a 1/3 titer or MOI 3.33 for each gRNA/CRISPR vector for tatABC and revABC, and a 1/6 titer or MOI 1.67 for each vector for tatrevABC. ELISA measurements clearly showed that p24 production was significantly lower in all single CRISPR-transduced cells than in WT (P = 0.001, 0.001, 0.001, 0.003, 0.001, and 0.003 for tatA, tatB, tatC, revA, revB, and revC, respectively), with the highest efficacy of a 48-fold reduction being observed in revB and no significant reduction being noted between empty LC-transduced cells and WT (P = 0.932). Multiple gRNA treatments resulted in similar or even lower p24 levels than those of single gRNAs (P = 0.003, 0.002, and 0.002 for tatABC, revABC, and tatrevABC, respectively, versus WT).
Latent HIV-1 infection in resting CD4+ T cells is the primary cause of the barrier to a functional cure. In order to establish whether our CRISPR constructs reached this cell population, we used J-Lat 10.6, a Jurkat CD4+ T-cell clone latently infected with the full-length R7 strain of HIV-1 minus env and nef13. HIV-1 transcription was silent in these cells due to a modified chromatin structure that prevents the access of the cellular transcriptional machinery to the HIV-1 LTR promoter. The treatment of cells with cytokines, such as TNFα, will induce chromatin remodeling, providing access to the HIV-1 promoter, its activation, and the initiation of viral protein production. J-Lat cells were transduced with single and multiple gRNA/CRISPR for five days similar to previous treatments on L-2 cells, the culture was changed to fresh medium, stimulated with 20 ng/mL TNFα, and p24 levels in the supernatant were measured 24 hours after induction (Fig. 4B). ELISA results showed that p24 production after reactivation was significantly lower in CRISPR-transduced J-Lat cells than in WT (P = 0.003, 0.003, 0.008, 0.01, 0.005, and 0.024 for tatA, tatB, tatC, revA, revB, and revC, respectively), with the highest efficacy of a 53-fold reduction being observed in tatA and no significant difference being noted between WT and empty LC-transduced cells (P = 0.132). Multiplex gRNA treatments showed similarly low or even lower p24 levels (P = 0.003, 0.003, and 0.003 for tatABC, revABC, and tatrevABC, respectively, versus WT). These results demonstrated that the lentiviral delivery of CRISPR/Cas9 successfully reached the isolated proviral genome, cleaved regulatory genes, and significantly inhibited viral replication, even after latency reversal.
Multiplexing gRNA/CRISPR constructs opens up the possibility of genomic excision occurring between two gRNA sites. In order to confirm this, PCR amplification spanning 2876 bp from upstream of the tat first exon to the end of the rev second exon was performed using DNA extracted from tatrevABC-transduced L-2 and J-Lat cells. DNA from WT and empty-LC transduced L-2 and J-Lat cells, as well as human T cell leukemia MT-4 cells as a mock sample, were amplified (Fig. 5C). Since all samples, excluding the mock, showed similar ~3000 bp amplicons, it was concluded that no excision was found between the tat first exon to rev second exon. The sequencing of amplicons further showed that mutations occurred at gRNA sites, albeit without any large excision between tat and rev. However, two samples showed smaller scale excisions: a 114-bp excision between tatA and tatC sites and a 54-bp excision between revB and revC in J-Lat and L-2 cells, respectively. Ultimately, concurrent mutations in multiple gRNA sites were evident in tatrevABC-transduced cells, which highlights the advantage of multiplexing gRNA to increase gene-editing efficiency.
Efficiency of the CRISPR/Cas9 construct in persistent and latent HIV-1 infection. (A) Persistently HIV-1-infected CD4+ T cells (L-2) were CRISPR-transduced with or without tat/rev-targeting gRNAs, either a single or multiple of three (tatABC and revABC) or six (tatrevABC). Transduction was performed for 5 days with MOI 10, the culture was changed to fresh medium, and p24 levels in the supernatant were measured 3 days later. A one-way ANOVA applied to all groups gave a p value < 0.001 and pairwise comparisons between WT and all other groups are shown. Assays were performed three times and average values (±SE) are shown. (B) Latently HIV-1-infected Jurkat T cells (J-Lat 10.6) were CRISPR-transduced with or without gRNAs similar to the treatment of L-2 cells. Transduction was performed for 5 days with MOI 10, the culture was changed to fresh medium, treated with or without TNFα 20 ng/mL, and p24 levels in the supernatant were measured 24 hours later. A one-way ANOVA applied to all groups gave a p value < 0.001 and pairwise comparisons between WT and all other groups are shown. Assays were performed three times and average values ±SE are shown. (C) Multiplexing gRNA failed to show excision between tat and rev. PCR amplification spanning 2876 bp between upstream of the tat first exon to the end of the rev second exon was conducted using DNA extracted from tatrevABC-transduced L-2 and J-Lat cells. Assays were performed three times and representative gel image are shown. The image was not joined from different parts of the gel. (D) Amplicons were Sanger sequenced and on-target gRNA sites from WT, empty LC, and several tatrevABC-transduced J-Lat and L-2 clones are shown. Wild-type NL4-3 sequences are shown topmost with the PAM site (NGG) indicated in blue. Mutations are indicated in red and Cas9 cleavage sites are indicated in red vertical lines.
Discussion
In the present study, we demonstrated that tat and rev-targeting CRISPR/Cas9 lentiviral constructs successfully abolished regulatory protein expression and function, and inhibited HIV-1 replication in persistently infected CD4+ T-cell lines as well as latently infected T cells. The regulatory proteins Tat and Rev play a crucial role in HIV-1 transcription and latency maintenance, and their genomes are among the most functionally conserved across subtypes, making them a compelling site of attack to suppress HIV-1. Our design selection of highly conserved tat and rev sequences was based on the HXB2 reference strain, and throughout this study, the constructs were shown to be effective against the HIV-1 NL4-3 strain integrated into 293 T and HeLa cells, the LAV strain inside L-2 cells, and the R7 strain inside J-Lat cells.
Previous studies suggested that CRISPR cleavage inside HIV-1 LTR cut out the whole length of the HIV-1 proviral genome by inducing DSB at both ends25,43. However, subsequent studies44 showed that sequencing analyses after LTR cleavage failed to reveal this excision and, thus, genome editing inside integral genes of HIV-1 may be more efficient to inhibit replication. The results of multiplexing gRNAs also failed to show any excision between tat and rev, presumably due to the host DNA repair mechanism after CRISPR-induced mutations. Other studies tested CRISPR attacks at the RRE inside the env gene to suppress nuclear export44; however, a secondary structure instead of the primary sequence is needed in order to maintain the Rev-RNA interaction, and base variations among RRE elements may only have subtle effects on Rev responsiveness45; therefore, targeting rev directly may be more efficient to achieve this purpose. One study also found that CRISPR/Cas9 transfection targeting the second exon of rev, at a site close to revA gRNA tested in this study, exhibited the highest degree of mutation out of LTR-, pol-, and rev-targeting gRNAs46.
In the present study, we demonstrated that revB and tatA gRNAs achieved the strongest HIV-1 suppression in persistently and latently infected T cells, respectively. tatB and revA were initially predicted to exert the highest replication inhibition due to the perceived importance of the targeted motifs for protein function47,48. However, other domains, such as the tat N-terminal acidic and rev multimerization domains, targeted by tatA and revB are also regarded as integral motifs49,50, and other factors, including CRISPR efficiency and mutation characteristics, may eventually play a larger role in influencing inhibition outcomes. Although sequencing showed an overall high frequency of mutations, it currently remains unclear whether the numbers of unique mutations, types of indels, or rates of frameshift mutations are mainly responsible for the varying degrees of efficiency between different gRNAs. The on-target analysis conducted in the present study was limited to Sanger sequencing, and the next generation sequencing of amplicons may provide a deeper understanding of mutation profiles and their relationship with CRISPR efficiency. However, replication inhibition was the most efficient by combining six tat- and rev-CRISPR gRNAs in a multiplex gRNA lentiviral cocktail.
Our CRISPR construct also showed efficacy in latently infected J-Lat 10.6 cells, proving that the lentiviral delivery of the CRISPR/Cas9 system successfully reached and cleaved the dormant HIV-1 proviral genome. The inaccessibility of transcriptionally inert HIV-1 DNA due to a modified chromatin structure was previously considered to inhibit gene editing, and a “shock and kill” approach using latency-reversing agents may be necessary16. However, the results of the present study suggest that CRISPR has the ability to target HIV-1 DNA without prior reactivation. The proviral genome found in latent infection is mostly defective51, whereas latently infected cell populations harboring intact tat/rev regions are capable of producing the virus upon the reversal of latency52, and targeting these genes to suppress reactivation is of significant value. Replication inhibition for several gRNAs, particularly rev-CRISPR, is weaker than that observed in persistently infected L-2 cells, which may prove the difficulty of genomic access to some extent. Another study also described a successful CRISPR attack on unstimulated latently HIV-1-infected J-Lat cells, albeit via nucleotransfection46. The capability of lentiviral vectors to transduce non-dividing cells, including resting CD4+ T cells, and maintain stable, long-term Cas9 transgene expression supports their potential use in eradicating infected cells constituting the latent reservoir. Cas9 expression was found to vary to some extent (Fig. 2A), but did not appear to correlate with CRISPR efficiency, which may further highlight the importance of the gRNA sequence. It is important to note that clonal cell lines were used in the present study, in which HIV-1 was integrated within specific sites in cellular DNA, and it currently remains unclear whether CRISPR efficiency is affected by differences in integration sites inside host DNA loci. However, our results showed viral suppression in both the latent state, in which HIV-1 DNA is frequently integrated in or near heterochromatin, and the persistent, transcriptionally active state, in which integration in the nuclear periphery is disfavored13,53.
The CRISPR recognition mechanism may tolerate certain mismatches to a varying degree and produce off-target mutagenesis, and the selection of a highly specific gRNA design is important for minimizing this effect. In the present study, simple Surveyor nuclease screening was used to show the absence of mutations in two of the most probable off-target sites of each gRNA inside the human genome. However, the Surveyor assay only serves as a preliminary mutation screening and may miss some mismatches in a heteroduplex DNA54. Assay results heavily depend on the quality of PCR amplicons and band intensities may underestimate the actual mutation events, as shown in the on-target lanes in Fig. 4. A comprehensive evaluation of off-target mutations may require deep whole-genome sequencing. Insertional mutagenesis potentially caused by a lentivirus may also cause genotoxicity and oncogenesis55; however, the cell viability assay performed in this study showed similar growth between WT and lentivirus-transduced cells.
Latent HIV-1 is a static sequence and non-evolving, with the lack of reverse transcription generally indicating that resistance-associated mutations do not occur56. However, similar to its capacity to escape from antiretroviral monotherapy and other gene editors, e.g. ZFN57, several novel findings have shown that mutations by NHEJ repair or a silent mutation occurring at the PAM recognition site may induce resistance to subsequent CRISPR attacks58,59. However, more recent studies discovered that the combination of two gRNAs circumvented this issue because repeated Cas9 cleavage may have led to the saturation of target mutations and prevented viral escape, analogous to the combination ART strategy. Dual gRNA combinations, particularly those targeting conserved and essential viral gene sequences, are able to mutate a large fraction of the provirus and inactivate it, creating major bottlenecks in viral evolution by severely limiting replication-competent virus variants60. In the present study, we combined even more gRNAs; six tat and rev gRNA combinations showed the similar or even stronger inhibition of replication than single gRNA. Viral escape is mainly facilitated by mutations that are non-deleterious to HIV-1 and, thus, the targeting of integral regulatory genes may minimize this effect because tat/rev dysfunctions inherently inactivate transcription and are primarily responsible for post-integration latency61. However, a viral resistance analysis was not performed in this study and repeated HIV-1 infections to CRISPR-transduced cells may shed further light on the emergence of tat and rev gRNA-specific escape mutants.
In conclusion, the results of the present study showed that the CRISPR/Cas9 system is robust and efficient for targeting the HIV-1 proviral genome to suppress replication in latency models. CRISPR efficiency mainly depends on how well the gRNA sequence matches the target DNA, and targeting of the highly conserved regulatory genes tat and rev is beneficial for this effect. Moreover, the importance of these genes to the transcription ability of HIV-1 may lessen the emergence of CRISPR escape mutants. The combination of multiple gRNAs may also maximize efficiency and minimize the risk of resistance. Recent studies may have proven efficacy in cell cultures and ex vivo; however, when in vivo safety and efficacy profiles have been established, CRISPR-bearing lentiviral vectors may be delivered into HIV-1-infected individuals to clear latent viral reservoirs. Based on the rapid advances being achieved in CRISPR/Cas9 research, a HIV-1 functional cure may soon be within reach.
Methods
gRNA designs and plasmids
Target sites from inside the tat and rev genes of the HIV-1 reference strain HXB2 genome (GenBank accession no. K30455), either the sense or antisense strand, were identified using the CRISPR design tool at http://crispr.mit.edu 62. Homology across subtypes was obtained by aligning the designed gRNAs to 97 HIV-1 strains from the Los Alamos National Lab (LANL) HIV Sequence Compendium using the QuickAlign v2 tool63, and percentage values signify the number of conserved nucleotides in the 23-bp on-target sequence. Nucleotide variability as measured by the average Shannon entropy score is also shown; a higher score indicates greater variability. Synthetic gRNA oligonucleotides were cloned into pLentiCRISPRv2 (Addgene #52961) at BsmBI restriction sites64.
Lentiviral vector construction and transduction
Lentiviral vectors were produced by co-transfecting 293 T cells with a cocktail of gRNA/Cas9-expressing lentiCRISPRv2, the Gag-Pol packaging plasmid psPAX2 (Addgene #12260), and VSVG expression vector pHIT/G65 at a ratio of 3:4:1 using FuGENE HD transfection reagent (Promega).
Transduction of the lentiviral vector into target cells was performed with MOI 10. MOI of 10 was selected because it was previously reported to be most optimal for expressing the transgene into target T cells with minimal cytotoxicity39. A preliminary analysis to assess the transducing unit (TU) was performed by transducing a GFP-bearing lentiviral vector in titration into 293 T cells and observing GFP expression under a fluorescent microscope in order to obtain TU/mL, before using it to calculate MOI. LentiCRISPRv2 without gRNA was used as an empty vector for mock transduction. Culture medium containing the virus was maintained for 5 days; second transduction was performed 7 days later, and the virus was also maintained for 5 days before proceeding with the analysis. Antibiotic selection was not performed after each transduction.
Multiplex gRNA transduction
Multiple gRNA transductions were performed by the co-infection of three tat and rev gRNA-bearing lentiviral vectors (tatABC and revABC) or six gRNAs (tatrevABC) in a lentiviral cocktail. MOI was the same in all multiplex gRNA groups as those in the single gRNA treatment groups. While MOI of 10 was used for single gRNA transduction, a 1/3 titer of each gRNA/CRISPR vector for tatABC and revABC, i.e. MOI 3.33 for each lentivirus, and a 1/6 titer of each vector for tatrevABC, i.e. MOI 1.67, for each lentivirus were used.
Cell cultures and development of stable Tat- and Rev-expressing cells
HEK 293 T and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS; BioWest). L-2 cells were grown in RPMI 1640 medium (Nacalai Tesque) with 10% FBS. J-Lat 10.6 cells were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, and were grown in RPMI 1640 medium with 10% FBS, 100 U/mL penicillin G, and 100 μg/mL streptomycin.
The tat and rev genomes were amplified from pNL4-366 and FLAG-tagged at the N- and C-terminal ends, respectively, before being cloned into the pLenti CMV Puro DEST backbone (Addgene #17452)67. Co-transfection into packaging 293 T cells was performed with psPAX2 and pHIT/G to produce VSVG-pseudotyped lentiviral vectors. Transduction into target 293 T and HeLa cells was performed with MOI of 1 and selected with puromycin. Limiting dilutions were performed and the strongest FLAG-expressing cells were selected by immunoblot assays.
Antibodies for Western blotting
Protein samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane, blocked with 5% non-fat milk in PBS, and immunostained with the primary antibody as follows: an anti-FLAG M2 monoclonal antibody (Sigma Aldrich) for FLAG expression, CRISPR/Cas9 polyclonal antibody (Epigentek) for Cas9, anti-HIV-1SF2 gp12068 for envelope gp120, and anti-beta-actin polyclonal antibody (BioVision) for beta-actin. Anti-HIV-1SF2 gp120 was obtained from NIH ARRRP. Primary antibody binding was followed with an incubation with peroxidase-labeled secondary antibodies and an immunocomplex was visualized with Pierce western blotting substrate (Thermo Scientific) using the OptimaShot CL-420α chemiluminescence imaging system (Wako). Blot images were cropped using Photoshop CS6 and any contrast adjustment was applied equally across the entire image, including control lanes.
Tat and Rev functional analysis
A Tat functional analysis was performed by transfecting target cells with pLTR-Luc, and three days after transfection, cells were lysed to measure luciferase activity. The plasmid pLTR-Luc contains HIV-1 3′LTR upstream of the luc gene and transactivation by Tat will drive luciferase expression69. Transfections of the mammalian expression vector pRc/RSV (RDB No 580, Riken Gene Bank) and the Tat expression vector pRSV-Tat70 into 293 T or HeLa cells were used as negative and positive controls, respectively.
A Rev functional analysis was performed by transfecting target cells with the gp140-expressing plasmid, and three days after transfection, the cell lysate was immunostained with an anti-gp120 antibody. The plasmid gp140 was constructed from wild-type gp160, which was terminated at the N terminus to the membrane-spanning region, had a serine substitution at the gp120-gp41 junction, and attached with the T4 bacteriophage fibritin at the C terminus in order to stabilize uncleaved gp140 trimers71. Transfections of pcDNA3.1/Zeo(+) (V86020, Invitrogen) and the Rev expression plasmid pCA-Rev72 into 293 T cells were used as negative and positive controls, respectively.
Reactivation for latent infection
Cytokine reactivation for latently infected J-Lat cells was performed by adding 20 ng/mL of TNFα into cells with fresh medium, which was conducted five days after the gRNA/CRISPR treatment. Twenty-four hours after reactivation, the supernatant was cleared from cell debris by centrifugation at 3000 RPM for 5 minutes, and p24 inside the supernatant was measured as described below.
p24 ELISA
HIV-1 p24 was quantified using anti-p24 ELISA (ZeptoMetrix RETROtek HIV-1 p24 antigen ELISA, Fisher Scientific) according to the manufacturer’s protocol. The timing of the p24 measurement was 3 days after the fresh medium change for L-2 cells and 24 hours after cytokine reactivation for J-Lat cells.
On- and off-target analyses
Genomic DNA was extracted from CRISPR-transduced and control cells 7 days after transduction using the QIAamp DNA Blood Minikit (Qiagen) before being used as templates for on- and off-target PCR. On-target amplicons were sequenced (Macrogen Inc.) and analyzed using GENETYX software ver. 10 (GENETYX Corp.).
Potential off-target sites inside the coding region of the human genome were identified using the aforementioned CRISPR design tool62. Two sites with the highest off-target probability from each gRNA were selected. PCR primers were designed to amplify 400–800-bp sequences from the human genome that contained each off-target site (Supplementary Table 1). Amplicons were hybridized before being digested with nuclease from the Surveyor mutation detection kit (IDT). Products were then analyzed on a 2% agarose gel, stained with GelRed (Biotium), and photographed.
Statistical analysis
All experiments in the present study were conducted with at least 3 replicates and data were shown as means ± standard deviations (SD). A one-way analysis of variance (ANOVA) was performed for samples from more than 2 groups, and pairwise comparisons using an independent-sample t-test were conducted if the ANOVA test gave a p value < 0.05. All analyses were performed with SPSS 23.0 (SPSS, Inc.).
Data availability
All necessary data generated or analyzed during the present study are included in this published article and its Supplementary Information files.
References
UNAIDS. Global AIDS Update 2016. (Joint United Nations Programme on HIV/AIDS, Geneva, 2016).
Maartens, G., Celum, C. & Lewin, S. R. HIV infection: epidemiology, pathogenesis, treatment, and prevention. The Lancet 384, 258–271 (2014).
Schrager, L. K. & D’Souza, M. P. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA 280, 67–71 (1998).
Finzi, D. et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997).
Chun, T. W. et al. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195, 1762–1764 (2007).
The International AIDS Society Scientific Working Group on HIV Cure. Towards an HIV cure: a global scientific strategy. Nat. Rev. Immunol. 12, 607–614 (2012).
Chomont, N. et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900 (2009).
Chun, T.-W. et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197, 714–720 (2008).
Montessori, V., Press, N., Harris, M., Akagi, L. & Montaner, J. S. Adverse effects of antiretroviral therapy for HIV infection. Can. Med. Assoc. J. 170, 229–238 (2004).
García, dO. P. et al. Impact of adherence and highly active antiretroviral therapy on survival in HIV-infected patients. J. Acquir. Immune Defic. Syndr. 30, 105–110 (2002).
Stadeli, K. M. & Richman, D. D. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir. Ther. 18, 115 (2013).
Sterne, J. A. et al. Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: a prospective cohort study. The Lancet 366, 378–384 (2005).
Jordan, A., Bisgrove, D. & Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 (2003).
Rasmussen, T. A. et al. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum. Vaccin. Immunother. 9, 993–1001 (2013).
Bullen, C. K., Laird, G. M., Durand, C. M., Siliciano, J. D. & Siliciano, R. F. Novel ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 (2014).
Shan, L. et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501 (2012).
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Joung, J. K. & Sander, J. D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Gaj, T., Gersbach, C. A. & Barbas, C. F. ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Nemudryi, A. A., Valetdinova, K. R., Medvedev, S. P. & Zakian, S. M. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae 6, 19–40 (2014).
Schumann, K. et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA 112, 10437–10442 (2015).
Weber, N. D., Aubert, M., Dang, C. H., Stone, D. & Jerome, K. R. DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward. Virology 454, 353–361 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Ebina, H., Misawa, N., Kanemura, Y. & Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510 (2013).
Emerman, M. & Malim, M. H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280, 1880–1884 (1998).
Cullen, B. R. & Greene, W. C. Regulatory pathways governing HIV-1 replication. Cell 58, 423–426 (1989).
Karn, J. & Stoltzfus, C. M. Transcriptional and posttranscriptional regulation of HIV-1gene expression. Cold Spring Harb. Perspect. Med. 2, a006916 (2012).
Hauber, J., Malim, M. H. & Cullen, B. R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J. Virol. 63, 1181–1187 (1989).
DiGiusto, D. L. et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2, 36ra43–36ra43 (2010).
Anderson, J. et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes—CCR5 ribozyme, tat-rev siRNA, and TAR decoy—in SCID-hu mouse–derived T cells. Mol. Ther. 15, 1182–1188 (2007).
Asparuhova, M. B. et al. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J. Gene Med. 9, 323–334 (2007).
Lo, K. S., Biasolo, M. A., Dehni, G., Palú, G. & Haseltine, W. A. Inhibition of replication of HIV-1 by retroviral vectors expressing tat-antisense and anti-tat ribozyme RNA. Virology 190, 176–183 (1992).
Michienzi, A., Li, S., Zaia, J. A. & Rossi, J. J. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl. Acad. Sci. USA 99, 14047–14052 (2002).
Lee, S.-W., Gallardo, H. F., Gilboa, E. & Smith, C. Inhibition of human immunodeficiency virus type 1 in human T cells by a potent Rev response element decoy consisting of the 13-nucleotide minimal Rev-binding domain. J. Virol. 68, 8254–8264 (1994).
Bevec, D., Dobrovnik, M., Hauber, J. & Böhnlein, E. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc. Natl. Acad. Sci. USA 89, 9870–9874 (1992).
Unwalla, H. & Banerjea, A. C. Inhibition of HIV-1 gene expression by novel macrophage-tropic DNA enzymes targeted to cleave HIV-1 TAT/Rev RNA. Biochem. J. 357, 147–155 (2001).
Chen, Z. et al. Inhibition of HIV-1 in human T-lymphocytes by retrovirally transduced anti-tat and rev hammerhead ribozymes. Gene 149, 33–39 (1994).
Jones, S. et al. Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum. Gene Ther. 20, 630–640 (2009).
Canver, M. C. et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 289, 21312–21324 (2014).
Fu, Y. et al. High frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Ohki, K. et al. Noninfectious doughnut-shaped human immunodeficiency virus type 1 can induce syncytia mediated by fusion of the particles with CD4-positive cells. J. Acquir. Immune Defic. Syndr. 4, 1233–1240 (1991).
Kaminski, R. et al. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 6, 22555 (2016).
Yoder, K. E. & Bundschuh, R. Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci. Rep. 6, 29530 (2016).
Olsen, H. S., Nelbock, P., Cochrane, A. W. & Rosen, C. A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science 247, 845–848 (1990).
Zhu, W. et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 12, 22 (2015).
Boykins, R. A. et al. Cutting edge: a short polypeptide domain of HIV-1-Tat protein mediates pathogenesis. J. Immunol. 163, 15–20 (1999).
Daugherty, M. D., Liu, B. & Frankel, A. D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 17, 1337–1342 (2010).
Rappaport, J., Lee, S. J., Khalili, K. & Wong-Staal, F. The acidic amino-terminal region of the HIV-1 Tat protein constitutes an essential activating domain. New Biol. 1, 101–110 (1989).
Thomas, S. L. et al. Functional analysis of the human immunodeficiency virus type1 Rev protein oligomerization interface. J. Virol. 72, 2935–2944 (1998).
Hiener, B. et al. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep. 21, 813–822 (2017).
Procopio, F. A. et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2, 874–883 (2015).
Dieudonné, M. et al. Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery. EMBO J. 28, 2231–2243 (2009).
Yeung, A. T., Hattangadi, D., Blakesley, L. & Nicolas, E. Enzymatic mutation detection technologies. Biotechniques 38, 749–758 (2005).
Schambach, A., Zychlinski, D., Ehrnstroem, B. & Baum, C. Biosafety features of lentiviral vectors. Hum. Gene Ther. 24, 132–142 (2013).
Siliciano, J. D. & Siliciano, R. F. A long-term latent reservoir for HIV-1: discovery and clinical implications. J. Antimicrob. Chemother. 54, 6–9 (2004).
De Silva Feelixge, H. S. et al. Detection of treatment-resistant infectious HIV after genome-directed antiviral endonuclease therapy. Antiviral Res. 126, 90–98 (2016).
Liang, C., Wainberg, M. A., Das, A. T. & Berkhout, B. CRISPR/Cas9: a double-edged sword when used to combat HIV infection. Retrovirology 13, 37 (2016).
Ueda, S., Ebina, H., Kanemura, Y., Misawa, N. & Koyanagi, Y. Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiol. Immunol. 60, 483–496 (2016).
Wang, G., Zhao, N., Berkhout, B. & Das, A. T. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep. 17, 2819–2826 (2016).
Emiliani, S. et al. Mutations in the tat gene are responsible for Human Immunodeficiency Virus type 1 postintegration latency in the U1 cell line. J. Virol. 72, 1666–1670 (1998).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Foley, B. et al. HIV Sequence Compendium 2014. (Los Alamos National Laboratory, Theoretical Biology and Biophysics, 2014).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Fouchier, R. A., Meyer, B. E., Simon, J. H., Fischer, U. & Malim, M. H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16, 4531–4539 (1997).
Adachi, A. et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291 (1986).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 (2009).
Haigwood, N. L. et al. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res. Hum. Retroviruses 6, 855–869 (1990).
Kameoka, M., Tanaka, Y., Ota, K., Itaya, A. & Yoshihara, K. Poly (ADP-ribose) polymerase is involved in PMA-induced activation of HIV-1 in U1 cells by modulating the LTR function. Biochem. Biophys. Res. Commun. 262, 285–289 (1999).
Kameoka, M. et al. Short communication: RNA interference directed against Axin1 upregulates human immunodeficiency virus type 1 gene expression by activating the Wnt signaling pathway in HeLa-derived J111 cells. AIDS Res. Hum. Retroviruses 25, 1005–1011 (2009).
Yang, X. et al. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76, 4634–4642 (2002).
Iwabu, Y. et al. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem. 284, 35060–35072 (2009).
Acknowledgements
We would like to thank K. Tokunaga (Department of Pathology, National Institute of Infectious Diseases, Tokyo, Japan) for the gift of pCA-Rev. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: (1) J-Lat Full Length Clone (clone# 9849) from Dr. Eric Verdin, (2) antiserum to HIV-1SF2 gp120 from Chiron Corporation. This work was supported in part by Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS); and the program of the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from the Ministry of Education, Culture, Sport, Science and Technology in Japan, and the Japan Agency for Medical Research and Development (AMED). The manuscript was proofread by Medical English Service, Kyoto, Japan.
Author information
Authors and Affiliations
Contributions
The study was conceived by Y.O. and M.K. Experiments and data analyses were performed by Y.O., M.I., and M.K. The manuscript was prepared by Y.O., T.K., and M.K. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ophinni, Y., Inoue, M., Kotaki, T. et al. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci Rep 8, 7784 (2018). https://doi.org/10.1038/s41598-018-26190-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26190-1
This article is cited by
-
NUMB facilitates autophagy initiation through targeting SCFβ-TrCP2 complex
Cell Death & Differentiation (2022)
-
Gene editing and RNAi approaches for COVID-19 diagnostics and therapeutics
Gene Therapy (2021)
-
CRISPR/Cas-New Molecular Scissors in Diagnostics and Therapeutics of COVID-19
Indian Journal of Clinical Biochemistry (2021)
-
An insight on promising strategies hoping to cure HIV-1 infection by targeting Rev protein—short review
Pharmacological Reports (2021)
-
Immunotherapy and vaccination against infectious diseases
Wiener klinische Wochenschrift (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.