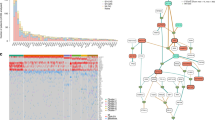
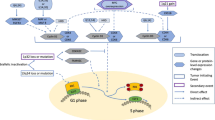
Key Points
-
The development of multiple myeloma is preceded by pre-malignant stages, and therefore constitutes a well-defined model of disease progression that is appropriate for studies of clonal evolution and heterogeneity
-
Whole-exome sequencing studies have enabled the characterization of the genomic alterations underlying the pathogenesis of multiple myeloma
-
The primary genomic events involved in multiple myeloma are the acquisition of hyperdiploidy or translocations affecting the IGH genes; these events are mutually exclusive
-
Secondary genomic events include chromosomal translocations, copy-number variations and single-nucleotide variants
-
Genomic events underlying multiple myeloma affect multiple signalling pathways including the MYC, NF-κB, and MAPK pathways, plasma-cell differentiation, cell-cycle regulation or DNA-damage repair
Abstract
Multiple myeloma (MM) is a genetically complex disease that evolves from pre-malignant stages, such as monoclonal gammaopathy of undetermined significance and smouldering multiple myeloma, and progresses to symptomatic MM; this continuum provides a unique framework to study the sequential genomic evolution of MM. In the past 5 years, results from large-scale whole-exome sequencing studies have provided new insights into the clonal heterogeneity and evolution of the disease. Moreover, the recurrent co-occurrence of genomic events helps to dissect the genomic complexity underlying tumour progression. According to the primary genetic events involved in tumorigenesis, MM tumours are hierarchically subdivided into hyperdiploid and non-hyperdiploid subtypes; subsequently, secondary genetic events lead to tumour progression. In this Review, we describe the 'driver' gene alterations involved in the development and progression of MM, with a focus on the sequential acquisition of the main genomic aberrations. We also provide valuable insight into the clonal heterogeneity and clonal evolution of the disease, as well as into the therapeutic implications of a comprehensive understanding of the genomic complexity of MM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
24 August 2016
In Figure 4, in the MYC activation subpanel, the genes depicted should be MYC and MAX instead of BIRC2 and NRAS. This error has been corrected in the print and online versions of the article.
References
Palumbo, A. & Anderson, K. Multiple myeloma. N. Engl. J. Med. 364, 1046–1060 (2011).
Howlader, N. et al. SEER Cancer Statistics Review, 1975–2013. SEER website [online] http://seer.cancer.gov/csr/1975_2013/ (2016).
Landgren, O. et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 113, 5412–5417 (2009).
Weiss, B. M., Abadie, J., Verma, P., Howard, R. S. & Kuehl, W. M. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood 113, 5418–5422 (2009).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Lohr, J. G. et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25, 91–101 (2014).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Walker, B. A. et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nature Commun. 6, 6997 (2015).
Walker, B. A. et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol, http://dx.doi.org/10.1200/JCO.2014.59.1503 (2015).
Altieri, A., Chen, B., Bermejo, J. L., Castro, F. & Hemminki, K. Familial risks and temporal incidence trends of multiple myeloma. Eur. J. Cancer 42, 1661–1670 (2006).
Broderick, P. et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat. Genet. 44, 58–61 (2012).
Chubb, D. et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat. Genet. 45, 1221–1225 (2013).
Weinhold, N. et al. Inherited genetic susceptibility to monoclonal gammopathy of unknown significance. Blood 123, 2513–2517; quiz 2593, http://dx.doi.org/10.1182/blood-2013-10-532283 (2014).
Weinhold, N. et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat. Genet. 45, 522–525 (2013).
Johnson, D. C. et al. Genetic factors influencing the risk of multiple myeloma bone disease. Leukemia, http://dx.doi.org/10.1038/leu.2015.342 (2015).
Swaminathan, B. et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat. Commun. 6, 7213 (2015).
Ziv, E. et al. Genome-wide association study identifies variants at 16p13 associated with survival in multiple myeloma patients. Nat. Commun. 6, 7539 (2015).
Landgren, O. et al. Racial disparities in the prevalence of monoclonal gammopathies: a population-based study of 12,482 persons from the National Health and Nutritional Examination Survey. Leukemia 28, 1537–1542 (2014).
Gonzalez, D. et al. Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood 110, 3112–3121 (2007).
Bergsagel, P. L. et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005).
Ross, F. M. et al. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia 19, 1634–1642 (2005).
Kaufmann, H. et al. Both IGH translocations and chromosome 13q deletions are early events in monoclonal gammopathy of undetermined significance and do not evolve during transition to multiple myeloma. Leukemia 18, 1879–1882 (2004).
Fonseca, R. et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 100, 1417–1424 (2002).
Avet-Loiseau, H. et al. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe Francophone Myelome. Cancer Res. 59, 4546–4550 (1999).
Chesi, M. et al. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood 88, 674–681 (1996).
Tanguay, D. A. & Chiles, T. C. Regulation of the catalytic subunit (p34PSK-J3/cdk4) for the major D-type cyclin in mature B lymphocytes. J. Immunol. 156, 539–548 (1996).
Solvason, N. et al. Induction of cell cycle regulatory proteins in anti-immunoglobulin-stimulated mature B lymphocytes. J. Exp. Med. 184, 407–417 (1996).
Walker, B. A. et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 121, 3413–3419 (2013).
Zhan, F. et al. The molecular classification of multiple myeloma. Blood 108, 2020–2028 (2006).
Keats, J. J. et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 101, 1520–1529 (2003).
Fonseca, R. et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 101, 4569–4575 (2003).
Chang, H. et al. The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. Br. J. Haematol. 125, 64–68 (2004).
Avet-Loiseau, H. et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood 109, 3489–3495 (2007).
Chesi, M. et al. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92, 3025–3034 (1998).
Santra, M., Zhan, F., Tian, E., Barlogie, B. & Shaughnessy, J. Jr. A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood 101, 2374–2376 (2003).
Keats, J. J. et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 105, 4060–4069 (2005).
Martinez-Garcia, E. et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117, 211–220 (2011).
Pei, H. et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011).
Cappellen, D. et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nature Genet. 23, 18–20 (1999).
San Miguel, J. F. et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med. 359, 906–917 (2008).
Avet-Loiseau, H. et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J. Clin. Oncol. 28, 4630–4634 (2010).
Hanamura, I. et al. Ectopic expression of MAFB gene in human myeloma cells carrying (14;20)(q32;q11) chromosomal translocations. Jpn. J. Cancer Res. 92, 638–644 (2001).
Hurt, E. M. et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5, 191–199 (2004).
Avet-Loiseau, H. et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood 117, 2009–2011 (2011).
Prideaux, S. M., Conway O'Brien, E. & Chevassut, T. J. The genetic architecture of multiple myeloma. Adv. Hematol. 2014, 864058 (2014).
Shaughnessy, J. Jr et al. Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood 98, 217–223 (2001).
Mikhael, J. R. et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 88, 360–376 (2013).
Ross, F. M. et al. The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica 95, 1221–1225 (2010).
Avet-Loiseau, H. et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood 98, 3082–3086 (2001).
Weinhold, N. et al. Concomitant gain of 1q21 and MYC translocation define a poor prognostic subgroup of hyperdiploid multiple myeloma. Haematologica, http://dx.doi.org/10.3324/haematol.2015.136929 (2015).
Haradhvala, N. J. et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell 164, 538–549 (2016).
Affer, M. et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28, 1725–1735 (2014).
Walker, B. A. et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 4, e191 (2014).
Carrasco, D. R. et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 9, 313–325 (2006).
Walker, B. A. et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 116, e56–65 (2010).
Walker, B. A. et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 108, 1733–1743 (2006).
Annunziata, C. M. et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Keats, J. J. et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007).
Smadja, N. V. et al. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia 12, 960–969 (1998).
Debes-Marun, C. S. et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia 17, 427–436 (2003).
Chng, W. J. et al. A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood 106, 2156–2161 (2005).
Onodera, N., McCabe, N. R. & Rubin, C. M. Formation of a hyperdiploid karyotype in childhood acute lymphoblastic leukemia. Blood 80, 203–208 (1992).
Fonseca, R. et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood 102, 2562–2567 (2003).
Pawlyn, C. et al. Coexistent hyperdiploidy does not abrogate poor prognosis in myeloma with adverse cytogenetics and may precede IGH translocations. Blood 125, 831–840 (2015).
Chng, W. J. et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 67, 2982–2989 (2007).
Smadja, N. V. et al. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood 98, 2229–2238 (2001).
Chretien, M. L. et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood 126, 2713–2719 (2015).
Shaughnessy, J. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology 10 (Suppl. 1), 117–126, http://dx.doi.org/10.1080/10245330512331390140 (2005).
Sawyer, J. R., Tricot, G., Mattox, S., Jagannath, S. & Barlogie, B. Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 91, 1732–1741 (1998).
Sawyer, J. R. et al. Jumping translocations of 1q12 in multiple myeloma: a novel mechanism for deletion of 17p in cytogenetically defined high-risk disease. Blood 123, 2504–2512 (2014).
Fournier, A. et al. 1q12 chromosome translocations form aberrant heterochromatic foci associated with changes in nuclear architecture and gene expression in B cell lymphoma. EMBO Mol. Med. 2, 159–171 (2010).
Shi, L. et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 1, 22–33 (2010).
Hanamura, I. et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 108, 1724–1732 (2006).
Boyd, K. D. et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin. Cancer Res. 17, 7776–7784 (2011).
Chang, H. et al. Impact of genomic aberrations including chromosome 1 abnormalities on the outcome of patients with relapsed or refractory multiple myeloma treated with lenalidomide and dexamethasone. Leuk. Lymphoma 51, 2084–2091 (2010).
Avet-Loiseau, H. et al. Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. 27, 4585–4590 (2009).
Chang, H. et al. 1p21 deletions are strongly associated with 1q21 gains and are an independent adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Bone Marrow Transplant. 45, 117–121 (2010).
Fonseca, R. et al. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 15, 981–986 (2001).
Avet-Loiseau, H. et al. Monosomy 13 is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Intergroupe Francophone Myelome. Blood 94, 2583–2589 (1999).
Chiecchio, L. et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 20, 1610–1617 (2006).
Avet-Louseau, H. Daviet, A., Sauner, S., Bataille, R. & Intergroupe Francophone du Myélome. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br. J. Haematol. 111, 1116–1117 (2000).
Fonseca, R. et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia 23, 2210–2221 (2009).
Tricot, G. et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 86, 4250–4256 (1995).
Tiedemann, R. E. et al. Genetic aberrations and survival in plasma cell leukemia. Leukemia 22, 1044–1052 (2008).
Lode, L. et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 95, 1973–1976 (2010).
Drach, J. et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood 92, 802–809 (1998).
Palumbo, A. et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin. Oncol. 33, 2863–2869 (2015).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl Acad. Sci. USA 104, 20007–20012 (2007).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nature Genet. 45, 1134–1140 (2013).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Weber, A. M. & Ryan, A. J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 (2015).
Dziembowski, A., Lorentzen, E., Conti, E. & Seraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 (2007).
Schmid, M. & Jensen, T. H. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33, 501–510 (2008).
Weissbach, S. et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br. J. Haematol. 169, 57–70 (2015).
Tam, W. et al. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 107, 4090–4100 (2006).
Pasqualucci, L. et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J. Exp. Med. 203, 311–317 (2006).
Landau, D. A., Carter, S. L., Getz, G. & Wu, C. J. Clonal evolution in hematological malignancies and therapeutic implications. Leukemia 28, 34–43 (2014).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Bahlis, N. J. Darwinian evolution and tiding clones in multiple myeloma. Blood 120, 927–928 (2012).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Melchor, L. et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 28, 1705–1715 (2014).
Smadja, N. V. et al. Further cytogenetic characterization of multiple myeloma confirms that 14q32 translocations are a very rare event in hyperdiploid cases. Genes Chromosomes Cancer 38, 234–239 (2003).
Walker, B. A. et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 28, 384–390 (2014).
Greipp, P. R. et al. International staging system for multiple myeloma. J. Clin. Oncol. 23, 3412–3420 (2005).
Andrulis, M. et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov. 3, 862–869 (2013).
Poulikakos, P. I., Zhang, C., Bollag, G., Shokat, K. M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010).
Lonial, S. et al. Elotuzumab in combination with lenalidomide and low-dose dexamethasone in relapsed or refractory multiple myeloma. J. Clin. Oncol. 30, 1953–1959 (2012).
Jakubowiak, A. J. et al. Phase I trial of anti-CS1 monoclonal antibody elotuzumab in combination with bortezomib in the treatment of relapsed/refractory multiple myeloma. J. Clin. Oncol. 30, 1960–1965 (2012).
Lokhorst, H. M. et al. Targeting CD38 with Daratumumab Monotherapy in Multiple Myeloma. N. Engl. J. Med. 373, 1207–1219 (2015).
Lonial, S. et al. Daratumumab monotherapy in patients with treatment-refractory multiple myeloma (SIRIUS): an open-label, randomised, phase 2 trial. Lancet 387, 1551–1560 (2016).
Carpenter, R. O. et al. B-Cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin. Cancer Res. 19, 2048–2060 (2013).
Ramos, C. A. et al. Clinical responses with T lymphocytes targeting malignancy-associated kappa light chains. J. Clin. Invest. http://dx.doi.org/10.1172/JCI86000 (2016).
Lesokhin, A. M. et al. Nivolumab in Patients With Relapsed or Refractory Hematologic Malignancy: Preliminary Results of a Phase Ib Study. J. Clin. Oncol. http://dx.doi.org/10.1200/JCO.2015.65.9789 (2016).
Herbst, R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Janne, P. A. et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 14, 38–47 (2013).
Adjei, A. A. et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol. 26, 2139–2146 (2008).
Abdel-Wahab, O. et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov. 4, 538–545 (2014).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Guagnano, V. et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2, 1118–1133 (2012).
Gavine, P. R. et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056 (2012).
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Baughn, L. B. et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 66, 7661–7667 (2006).
Acknowledgements
We acknowledge funding from the Leukaemia & Lymphoma Society, the Multiple Myeloma Research Foundation and the US NIH.
Author information
Authors and Affiliations
Contributions
S.M., K.Z.S. and J.P. researched data for the article. S.M., D.A.L., G.G. and I.M.G. contributed to discussions of content. All authors were involved in writing the article, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Manier, S., Salem, K., Park, J. et al. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol 14, 100–113 (2017). https://doi.org/10.1038/nrclinonc.2016.122
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2016.122
This article is cited by
-
RNA processing mechanisms contribute to genome organization and stability in B cells
Oncogene (2024)
-
ARK5 enhances cell survival associated with mitochondrial morphological dynamics from fusion to fission in human multiple myeloma cells
Cell Death Discovery (2024)
-
Enhancing prognostic power in multiple myeloma using a plasma cell signature derived from single-cell RNA sequencing
Blood Cancer Journal (2024)
-
The role of next-generation sequencing in hematologic malignancies
Blood Research (2024)
-
Single-cell technologies in multiple myeloma: new insights into disease pathogenesis and translational implications
Biomarker Research (2023)