Abstract
Objective
To compare the clinical outcome of a multiplex polymerase chain reaction (PCR) based molecular diagnostic method -- Syndrome Evaluation System (SES) directed treatment strategy vs. standard of care (blood culture) directed treatment strategy for neonatal sepsis.
Methods
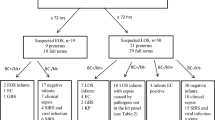
This randomized controlled trial (RCT) included 385 neonates with sepsis who were randomized into two groups -- SES and control (BACTEC). Both tests were performed for all the neonates. However, in the SES group, the results of SES test were revealed to the treating clinicians, while in the control group, SES results were withheld. Two ml of blood was drawn from each baby. One aliquot was sent for blood culture, whereas the remaining aliquot was sent for SES. Babies were then administered empirical IV antibiotics and given supportive care. Further antibiotic changes, if required were done in SES and control groups based on their respective reports. The microbiological profile, immediate outcome, duration of hospital stay, number of antibiotics used and readmission within a month in both groups were compared.
Results
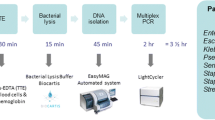
SES was better than BACTEC in identifying the causative organism in both the groups (68 % vs. 18 % in SES group and 72 % vs. 18 % in control group). SES had 100 % concordance with blood culture by BACTEC. Detection of bacteria and fungi were four and ten-fold higher respectively with SES when compared to BACTEC culture. Microbiological diagnosis was rapid with SES compared to BACTEC (7 h vs. 72 h). Treatment based on SES resulted in significantly less mortality (3 % vs. 18 %). Readmission rate, duration of hospital stay and change in antibiotics were also significantly less in SES group.
Conclusions
This new molecular based diagnostic system (SES) helps in rapid and accurate diagnosis of neonatal sepsis and reduces mortality and morbidity in affected neonates.



Similar content being viewed by others
References
Ostrosky-Zeichner L, Pappas PG. Invasive candidiasis in the intensive care unit. Crit Care Med. 2006;34:857–63.
Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immune modulating therapy for severe sepsis. Am J Med. 2003;115:529–35.
Read SJ, Jeffery KJ, Bangham CM. Aseptic meningitis and encephalitis: the role of PCR in the diagnostic laboratory. J Clin Microbiol. 1997;35:691–6.
Whitley RJ, Lakeman F. Herpes simplex virus infections of the central nervous system: therapeutic and diagnostic considerations. Clin Infect Dis. 1995;20:414–20.
Venkatesh M, Flores A, Luna RA, Versalovic J. Molecular microbiologcal methods in the diagnosis of neonatal sepsis. Expert Rev Anti Infect Ther. 2010;8:1037–48.
Ohlin A, Backman A, Bjorkqvist M, Molling P, Jurstrand M, Schollin J. Real-time PCR of the 16S-rRNA gene in the diagnosis of neonatal bacteremia. Acta Paediatr. 2008;97:1376–80.
Dierkes C, Ehrenstein B, Siebig S, Linde HJ, Reischl U, Salzberger B. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis. 2009;9:126.
Tirodker UH, Nataro JP, Smith S, LasCasas L, Fairchild KD. Detection of fungemia by polymerase chain reaction in critically ill neonates and children. J Perinatol. 2003;23:117–22.
Thomas LC, Gidding HF, Ginn AN, Olma T, Iredell J. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. J Microbiol Methods. 2007;68:296–302.
Enomoto M, Morioka I, Morisawa T, Yokoyama N, Matsuo M. A novel diagnostic tool for detecting neonatal infections using multiplex polymerase chain reaction. Neonatology. 2009;96:102–8.
European Medicine Agency. Report on the Expert Meeting on Neonatal and Paediatric Sepsis. EMA/477725/2010. http://www.ema.europa.eu/docs/en_GB/document_library/Report/2010/12/WC500100199.pdf. Accessed on November 3, 2015.
Stoll BJ, Hansen N, Fanaroff AA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–91.
Adams-Chapman I, Stoll BJ. Neonatal infection and long-term neurodevelopmental outcome in the preterm infant. Curr Opin Infect Dis. 2006;19:290–7.
Stoll BJ, Hansen NI, Adams-Chapman I, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004;292:2357–65.
Schelonka RL, Chai MK, Yoder BA, Hensley D, Brockett RM, Ascher DP. Volume of blood required to detect common neonatal pathogens. J Pediatr. 1996;129:275–8.
Forrest GN, Roghmann MC, Toombs LS, et al. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother. 2008;52:3558–63.
Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58:154–8.
Ly T, Gulia J, Pyrgos V, Waga M, Shoham S. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag. 2008;4:637–40.
Ohlin A, Bäckman A, Ewald U, Schollin J, Björkqvist M. Diagnosis of neonatal sepsis by broad-range 16S real-time polymerase chain reaction. Neonatology. 2012;101:241–6.
Kuruvilla KA, Swati P, Mary J, Jana KA. Bacterial profile in sepsis in a neonatal unit in South India. Indian Pediatr. 1998;35:851–8.
Shankar MJ, Agarwal R, Deorari AK, Paul VK. Sepsis in newborn. Indian J Pediatr. 2008;75:261–6.
Desai KJ, Malek SS, Parikh A. Neonatal septicemia: bacterial isolates and their antibiotic susceptibility patterns. Gujarat Med J. 2011;66:13–5.
Zaidi AKM, Thaver D, Ali SA, Khan TA. Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect Dis J. 2009;28:S10–8.
Khan SN, Joseph S. Neonatal sepsis: antibiotic sensitivity & resistance pattern of commonly isolated pathogens in a neonatal intensive care unit of a teritiary care hospital, South India. Int J Pharm Bio Sci. 2012;3:802–9.
Rathi MR, De AS, Mathur MM. Neonatal septicemia due to acinetobacter species and their antimicrobial susceptibility pattern. Ind Med Gaz. 2011;10:391–3.
Mustafa M, Ahmed SL. Bacteriological profile and antibiotic susceptibility patterns in neonatal septicemia in view of emerging drug resistance. J Med Allied Sci. 2014;4:02–8.
Jeyamurugan T, Ragulganesh R, Sucilathangam G, Ashihabegum MA, Velvizhi G, Palaniappan N. Acinetobacter spp.: an emerging pathogen in neonatal septicaemia. J Clin Diagn Res. 2012;6:805–6.
Mishra A, Mishra S, Jaganath G, Mittal RK, Gupta PK, Patra DP. Acinetobacter sepsis in newborns. Indian Pediatr. 1998;35:27–32.
Sharma CM, Agrawal RP, Sharan H, Kumar B, Sharma D, Bhatia SS. “Neonatal Sepsis”: bacteria & their susceptibility pattern towards antibiotics in neonatal intensive care unit. J Clin Diagn Res. 2013;7:2511–3.
Shete VB, Ghadage DP, Muley VA, Bhore AV. Acinetobacter septicemia in neonates admitted to intensive care units. J Lab Physicians. 2009;1:73–6.
De AS, Rathi MR, Mathur MM. Mortality audit of neonatal sepsis secondary to acinetobacter. J Glob Infect Dis. 2013;5:3–7.
Shah AJ, Mulla SA, Revdiwala SB. Neonatal sepsis: high antibiotic resistance of the bacterial pathogens in a neonatal intensive care unit of a tertiary care hospital. J Clin Neonatol. 2012;1:72–5.
Goel N, Ranjan PK, Aggarwal R, Chaudhary U, Sanjeev N. Emergence of nonalbicans candida in neonatal septicemia and antifungal susceptibility: experience from a tertiary care center. J Lab Physicians. 2009;1:53–5.
Sardana V, Pandey A, Madan M, Goel SP, Asthana AK. Neonatal candidemia: a changing trend. Indian J Pathol Microbiol. 2012;55:132–3.
Rani R, Mohapatra NP, Mehta G, Randhawa VS. Changing trends of candida species in neonatal septicaemia in a tertiary North Indian hospital. Indian J Med Microbiol. 2002;20:42–4.
Pammi M, Zhong D, Jhonson Y, Revell P, Versalovic J. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect Dis. 2014;14:390.
Tsai M-H, Chu S-M, Hsu J-F, et al. Polymicrobial bloodstream infection in neonates: microbiology, clinical characteristics, and risk factors. PLoS One. 2014;9:e830–82.
Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213.
Okeke IN, Peeling RW, Goossens H, et al. Diagnostics as essential tools for containing antibacterial resistance. Drug Resist Updat. 2011;14:95–106.
Dark PM, Dean P, Warhurst G. Bench-to-bedside review: the promise of rapid infection diagnosis during sepsis using polymerase chain reaction-based pathogen detection. Crit Care. 2009;13:217–9.
Wellinghausen N, Kochem AJ, Disqué C, et al. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol. 2009;47:2759–65.
Casalta JP, Gouriet F, Roux V, Thuny F, Habib G, Raoult D. Evaluation of the lightcycler septifast test in the rapid etiologic diagnostic of infectious endocarditis. Eur J Clin Microbiol Infect Dis. 2009;28:569–73.
Lehmann LE, Alvarez J. Potential clinical utility of polymerase chain reaction in microbiological testing for sepsis. Crit Care Med. 2009;37:3085–90.
Lehmann LE, Hunfeld KP. Improved detection of blood stream pathogens by real-time PCR in severe sepsis. Intensive Care Med. 2010;36:49–56.
Acknowledgments
The authors thank Mr. Dipanjan Chakraborty, Head of clinical studies, XCyton Diagnostics Pvt. Ltd for his inputs during the manuscript preparation.
Contributions
BVB, PP, BA and BS were the clinical investigators; BNH did the laboratory analysis; VBRK, KK, AR, and GM conducted the SES test. BVB will act as guarantor for this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors (Ravi Kumar BV, Krishnakumari K, Rekha A, Manjunath G) are affiliated to (employed by) XCyton Diagnostics Pvt. Ltd., which holds the patent/know how for Syndrome Evaluation System (SES). Hence, there is a potential conflict of interest.
Source of Funding
Council for Scientific and Industrial Research (CSIR), New Millennium Indian Technological Leadership Initiative.
Rights and permissions
About this article
Cite this article
Bhat, B.V., Prasad, P., Ravi Kumar, V.B. et al. Syndrome Evaluation System (SES) versus Blood Culture (BACTEC) in the Diagnosis and Management of Neonatal Sepsis - A Randomized Controlled Trial. Indian J Pediatr 83, 370–379 (2016). https://doi.org/10.1007/s12098-015-1956-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12098-015-1956-3