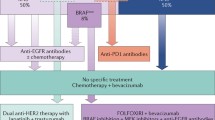
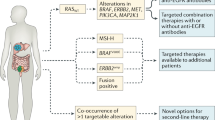
Opinion statement
The heterogenous nature of colorectal cancer (CRC) renders it a major clinical challenge. Increasing genomic understanding of CRC has improved our knowledge of this heterogeneity and the main cancer drivers, with significant improvements in clinical outcomes. Comprehensive molecular characterization has allowed clinicians a more precise range of treatment options based on biomarker selection. Furthermore, this deep molecular understanding likely extends therapeutic options to a larger number of patients. The biological associations of consensus molecular subtypes (CMS) with clinical outcomes in localized CRC have been validated in retrospective clinical trials. The prognostic role of CMS has also been confirmed in the metastatic setting, with CMS2 having the best prognosis, whereas CMS1 tumors are associated with a higher risk of progression and death after chemotherapy. Similarly, according to mesenchymal features and immunosuppressive molecules, CMS1 responds to immunotherapy, whereas CMS4 has a poorer prognosis, suggesting that a CMS1 signature could identify patients who may benefit from immune checkpoint inhibitors regardless of microsatellite instability (MSI) status. The main goal of these comprehensive analyses is to switch from “one marker-one drug” to “multi-marker drug combinations” allowing oncologists to give “the right drug to the right patient.” Despite the revealing data from transcriptomic analyses, the high rate of intra-tumoral heterogeneity across the different CMS subgroups limits its incorporation as a predictive biomarker. In clinical practice, when feasible, comprehensive genomic tests should be performed to identify potentially targetable alterations, particularly in RAS/BRAF wild-type, MSI, and right-sided tumors. Furthermore, CMS has not only been associated with clinical outcomes and specific tumor and patient phenotypes but also with specific microbiome patterns. Future steps will include the integration of clinical features, genomics, transcriptomics, and microbiota to select the most accurate biomarkers to identify optimal treatments, improving individual clinical outcomes. In summary, CMS is context specific, identifies a level of heterogeneity beyond standard genomic biomarkers, and offers a means of maximizing personalized therapy.
Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801.
Van Emburgh BO, Arena S, Siravegna G, Lazzari L, Crisafulli G, Corti G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:1–9.
Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–6.
Piskol R, Huw L, Sergin I, Kljin C, Modrusan Z, Kim D, et al. A clinically applicable gene-expression classifier reveals intrinsic and extrinsic contributions to consensus molecular subtypes in primary and metastatic colon cancer. Clin Cancer Res. 2019;25(14):4431–42. Using NanoString-based CMS classifier, the authors state that the two novel in vivo orthotopic implantation models reinforces the notion that extrinsic factors may impact on CMS identification in matched primary and metastatic colorectal cancer.
Alderdice M, Richman SD, Gollins S, Stewart JP, Hurt C, Adams R, et al. Prospective patient stratification into robust cancer-cell intrinsic subtypes from colorectal cancer biopsies. J Pathol. 2018;245(1):19–28.
Dunne PD, McArt DG, Bradley CA, O’Reilly PG, Barrett HL, Cummins R, et al. Challenging the cancer molecular stratification dogma: intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res. 2016;22(16):4095–104.
Laurent-Puig P, Marisa L, Ayadi M, Blum Y, Balogoun R, Pilati C, et al. Colon cancer molecular subtype intratumoral heterogeneity and its prognostic impact: an extensive molecular analysis of the PETACC-8. Ann Oncol. 2018;29:viii18.
Boeckx N, Koukakis R, Op de Beeck K, Rolfo C, Van Camp G, Siena S, et al. Primary tumor sidedness has an impact on prognosis and treatment outcome in metastatic colorectal cancer: results from two randomized first-line panitumumab studies. Ann Oncol. 2017;28(8):1862–8.
Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, et al. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular subtypes. Clin Cancer Res. 2018;24(5):1062–72.
Prior IA, Lewis PD, Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res. 2012;72(10):2457–67.
Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341(6239):209–14.
Milburn M, Tong L, DeVos A, Brunger A, Yamaizumi Z, Nishimura S, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science (80- ). 1990;247(4945):939–45.
Vetter IR. The guanine nucleotide-binding switch in three dimensions. Science (80- ). 2001;294(5545):1299–304.
Han C-B, Li F, Ma J-T, Zou H-W. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest. 2012;30(10):741–7.
Yokota T. Are KRAS/BRAF mutations potent prognostic and/or predictive biomarkers in colorectal cancers? Anticancer Agents Med Chem. 2012;12(2):163–71.
Schirripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer. 2015;136(1):83–90.
Wong NACS, Gonzalez D, Salto-Tellez M, Butler R, Diaz-Cano SJ, Ilyas M, et al. RAS testing of colorectal carcinoma—a guidance document from the Association of Clinical Pathologists Molecular Pathology and Diagnostics Group. J Clin Pathol. 2014;67(9):751–7.
Manuscript A, Malignancies H, Bettegowda C, Sausen M, Leary RJ, Kinde I, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28(6):1294–301. Plasma RAS determination showed high overall concordance with tissue and captured a mCRC population responsive to anti-EGFR therapy with the same predictive level as SoC tissue testing. The feasibility and practicality of ctDNA analysis may translate into an alternative tool for anti-EGFR treatment selection.
Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab–FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N Engl J Med. 2013;369(11):1023–34.
Cutsem E Van, Lenz H, Köhne C, Heinemann V, Tejpar S, Melezínek I. Fluorouracil , Leucovorin , and Irinotecan Plus Cetuximab treatment and RAS mutations in colorectal cancer. 2019;33(7).
Bokemeyer C, Ko C. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. 2015;1243–52.
Santos C, Azuara D, Viéitez JM, Páez D, Falcó E, Élez E, et al. Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: the ULTRA trial. Ann Oncol. 2019;30(5):796–803.
AACR Project GENIE. powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–31.
Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS G12C Inhibition with Sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–17. The first KRAS G12C inhibitor demonstrates clinical activity among different cancer types.
Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS G12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10(1):54–71.
Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRAS G12C inhibition in colorectal cancer. Cancer Discov. 2020;10(8):1129–39.
Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur J Cancer. 2015;51(5):587–94. This paper shown that BRAF mutated colorectal cancer didn’t achive significant clinical improvement with antiEGFR.
Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112(12):1888–94.
Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated reactivation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2(3):227–35.
Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–3.
Van Geel RMJM, Tabernero J, Elez E, Bendell JC, Spreafico A, Schuler M, et al. A phase Ib dose-escalation study of encorafenib and cetuximab with or without alpelisib in metastatic BRAF-mutant colorectal cancer. Cancer Discov. 2017;7(6):610–9.
Atreya CE, Van Cutsem E, Bendell JC, Andre T, Schellens JHM, Gordon MS, et al. Updated efficacy of the MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal cancer (mCRC). J Clin Oncol. 2015;33(15_suppl):103.
Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino T, et al. Encorafenib plus Cetuximab as a new standard of care for previously treated BRAF V600E–mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON Study. J Clin Oncol. 2021;39(4):273–84. Updated results from the practice changing BEACON clinical trial. This trial show that encorafenib-cetuximab has better OR, PFS and OS that irinotecan-based chemotherapy.
Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, et al. Non-V600 BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol. 2017;35(23):2624–30.
Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017;548:234.
Barras D, Missiaglia E, Wirapati P, Sieber OM, Jorissen RN, Love C, et al. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin Cancer Res. 2017;23(1):104–15.
Middleton G, Yang Y, Campbell CD, André T, Atreya CE, Schellens JHM, et al. BRAF-mutant transcriptional subtypes predict outcome of combined BRAF, MEK, and EGFR blockade with Dabrafenib, Trametinib, and Panitumumab in patients with colorectal cancer. Clin Cancer Res. 2020;26(11):2466–76.
Corcoran R, Giannakis M, Allen J, Chen J, Pelka K, Chao S, et al. SO-26 Clinical efficacy of combined BRAF, MEK, and PD-1 inhibition in BRAFV600E colorectal cancer patients. Ann Oncol. 2020;31(226–227):S226–7.
Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF V600E-mutant colorectal cancer. Cancer Discov. 2018;8(4):428–43.
Ross JS, Fakih M, Ali SM, Elvin JA, Schrock AB, Suh J, et al. Targeting HER2 in colorectal cancer: the landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 2018;124(7):1358–73.
Valtorta E, Martino C, Sartore-Bianchi A, Penaullt-Llorca F, Viale G, Risio M, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol. 2015;28(11):1481–91.
Loree JM, Bailey AM, Johnson AM, Yu Y, Wu W, Bristow CA, et al. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. JNCI J Natl Cancer Inst. 2018;110(12):1409–17.
Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient- derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1(6):508–23.
Raghav KPS, Overman MJ, Yu R, Meric-Bernstam F, Menter D, Kee BK, et al. HER2 amplification as a negative predictive biomarker for anti-epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. J Clin Oncol. 2016;34(15_suppl):3517.
Bregni G, Sciallero S, Sobrero A. HER2 amplification and anti-EGFR sensitivity in advanced colorectal cancer. JAMA Oncol. 2019;5(5):605.
Pietrantonio F, Vernieri C, Siravegna G, Mennitto A, Berenato R, Perrone F, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23(10):2414–22.
Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–46. The combination of trastuzumab and lapatinib is active and well tolerated in treatment-refractory patients with HER2-positive metastatic colorectal cancer.
Hainsworth JD, Meric-Bernstam F, Swanton C, Hurwitz H, Spigel DR, Sweeney C, et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: Results from mypathway, an open-label, phase IIA multiple basket study. J Clin Oncol. 2018;36(6):536–42.
Siravegna G, Sartore-Bianchi A, Nagy RJ, Raghav K, Odegaard JI, Lanman RB, et al. Plasma HER2 (ERBB2) copy number predicts response to HER2-targeted therapy in metastatic colorectal cancer. Clin Cancer Res. 2019;25(10):3046–53.
Nakamura Y, Okamoto W, Kato T, Hasegawa H, Kato K, Iwasa S, et al. TRIUMPH: Primary efficacy of a phase II trial of trastuzumab (T) and pertuzumab (P) in patients (pts) with metastatic colorectal cancer (mCRC) with HER2 (ERBB2) amplification (amp) in tumour tissue or circulating tumour DNA (ctDNA): A GOZILA sub-study. Ann Oncol. 2019;30:v199-200.
Strickler JH, Zemla T, Ou F-S, Cercek A, Wu C, Sanchez FA, et al. Trastuzumab and tucatinib for the treatment of HER2 amplified metastatic colorectal cancer (mCRC): Initial results from the MOUNTAINEER trial. Ann Oncol. 2019;30:v200.
Sartore-Bianchi A, Lonardi S, Martino C, Fenocchio E, Tosi F, Ghezzi S, et al. Pertuzumab and trastuzumab emtansine in patients with HER2-amplified metastatic colorectal cancer: the phase II HERACLES-B trial. ESMO open. 2020;5(5):e000911.
Li AY, McCusker MG, Russo A, Scilla KA, Gittens A, Arensmeyer K, et al. RET fusions in solid tumors. Cancer Treat Rev. 2019;81:101911.
Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Morano F, Fucà G, et al. RET fusions in a small subset of advanced colorectal cancers at risk of being neglected. Ann Oncol. 2018;29(6):1394–401.
Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6(1):7002.
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5(1):4846.
Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med. 2018;378(8):731–9.
Drilon A, Siena S, Ou S-HI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor Entrectinib: combined results from two phase i trials (ALKA-372–001 and STARTRK-1). Cancer Discov. 2017;7(4):400–9.
Sartore-Bianchi A, Ardini E, Bosotti R, Amatu A, Valtorta E, Somaschini A, et al. Sensitivity to Entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. JNCI J Natl Cancer Inst. 2016;108(1).
Demetri GD, Paz-Ares L, Farago AF, Liu S V, Chawla SP, Tosi D, et al. LBA17Efficacy and safety of entrectinib in patients with NTRK fusion-positive (NTRK-fp) tumors: pooled analysis of STARTRK-2, STARTRK-1 and ALKA-372–001. Ann Oncol. 2018;29(suppl_8).
Siena S, Doebele RC, Shaw AT, Karapetis CS, Tan DS-W, Cho BC, et al. Efficacy of entrectinib in patients (pts) with solid tumors and central nervous system (CNS) metastases: integrated analysis from three clinical trials. J Clin Oncol. 2019;37(15 suppl):3017.
Cortes-Ciriano I, Lee S, Park W-Y, Kim T-M, Park PJ. ARTICLE A molecular portrait of microsatellite instability across multiple cancers. 2017.
Ma J, Setton J, Lee NY, Riaz N, Powell SN. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat Commun. 2018;9(1):3292.
Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019.
Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–18.
Roth AD, Delorenzi M, Tejpar S, Yan P, Klingbiel D, Fiocca R, et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. JNCI J Natl Cancer Inst. 2012;104(21):1635–46.
Mohan HM, Ryan E, Balasubramanian I, Kennelly R, Geraghty R, Sclafani F, et al. Microsatellite instability is associated with reduced disease specific survival in stage III colon cancer. Eur J Surg Oncol. 2016;42(11):1680–6.
Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair Status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS Studies. Clin Cancer Res. 2014;20(20):5322–30.
Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–8.
Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, et al. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154(6):1805–13.
Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91(12):2417–22.
Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: Pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753–8.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–20.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13.
Le D, Kavan P, Kim T, Burge M, Van Cutsem E, Hara H, et al. O-021Safety and antitumor activity of pembrolizumab in patients with advanced microsatellite instability–high (MSI-H) colorectal cancer: KEYNOTE-164. Ann Oncol. 2018;29(suppl_5).
Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–91.
Andre T, Lonardi S, Wong M, Lenz H-J, Gelsomino F, Aglietta M, et al. Nivolumab + ipilimumab combination in patients with DNA mismatch repair-deficient/microsatellite instability-high (dMMR/MSI-H) metastatic colorectal cancer (mCRC): first report of the full cohort from CheckMate-142. J Clin Oncol. 2018;36(4_suppl):553–553.
André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–18. The first targeted treatment that demonstrate clinical activity in the first-line scenario. Pembrolizumab group has longer progression-free survival than chemotherapy group for MSI-H–dMMR metastatic colorectal cancer, with fewer treatment-related adverse events.
Andre T, Amonkar M, Norquist JM, Shiu K-K, Kim TW, Jensen BV, et al. Health-related quality of life in patients with microsatellite instability-high or mismatch repair deficient metastatic colorectal cancer treated with first-line pembrolizumab versus chemotherapy (KEYNOTE-177): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021.
Gong J, Wang C, Lee PP, Chu P, Fakih M. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring a POLE mutation. J Natl Compr Canc Netw. 2017;15(2):142–7.
Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1(3):207–16.
Tian S, Roepman P, Popovici V, Michaut M, Majewski I, Salazar R, et al. A robust genomic signature for the detection of colorectal cancer patients with microsatellite instability phenotype and high mutation frequency. J Pathol. 2012;228(4):586–95.
Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320–9.
De Sousa E, Melo F, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LPMH, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–8.
Trinh A, Trumpi K, De Sousa E, Melo F, Wang X, de Jong JH, Fessler E, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res. 2017;23(2):387–98.
Lan Y, Zhang D, Xu C, Hance KW, Marelli B, Qi J, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med. 2018;10(424).
Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–43.
Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590.
Janney A, Powrie F, Mann EH. Host–microbiota maladaptation in colorectal cancer. Nature. 2020;585(7826):509–17.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Lower Gastrointestinal Cancers
Rights and permissions
About this article
Cite this article
Ros, J., Baraibar, I., Martini, G. et al. The Evolving Role of Consensus Molecular Subtypes: a Step Beyond Inpatient Selection for Treatment of Colorectal Cancer. Curr. Treat. Options in Oncol. 22, 113 (2021). https://doi.org/10.1007/s11864-021-00913-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s11864-021-00913-5