Abstract
Background:
Enterococci are important pathogens in nosocomial infections. Various types of antibiotics, such as aminoglycosides, are used for treatment of these infections. Enterococci can acquire resistant traits, which can lead to therapeutic problems with aminoglycosides.Objectives:
This study was designed to identify the prevalence of, and to compare, the aac(6’)-aph(2”) and aph(3)-IIIa genes and their antimicrobial resistance patterns among Enterococcus faecalis and E. faecium isolates from patients at Imam Reza hospital in Kermanshah in 2011 - 2012.Patients and Methods:
One hundred thirty-eight clinical specimens collected from different wards of Imam Reza hospital were identified to the species level by biochemical tests. Antimicrobial susceptibility tests against kanamycin, teicoplanin, streptomycin, imipenem, ciprofloxacin, and ampicillin were performed by the disk diffusion method. The minimum inhibitory concentrations of gentamicin, streptomycin, kanamycin, and amikacin were evaluated with the microbroth dilution method. The aminoglycoside resistance genes aac(6’)-aph(2”) and aph(3”)-IIIa were analyzed with multiplex PCR.Results:
The prevalence of isolates was 33 (24.1%) for E. faecium and 63 (46%) for E. faecalis. Eighty-nine percent of the isolates were high-level gentamicin resistant (HLGR), and 32.8% of E. faecium isolates and 67.2% of E. faecalis isolates carried aac(6’)-aph(2”). The prevalence of aph(3”)-IIIa among the E. faecalis and E. faecium isolates was 22.7% and 77.3%, respectively.Conclusions:
Remarkably increased incidence of aac(6’)-aph(2”) among HLGR isolates explains the relationship between this gene and the high level of resistance to aminoglycosides. As the resistant gene among enterococci can be transferred, the use of new-generation antibiotics is necessary.Keywords
1. Background
Enterococci have changed from commensal intestine organisms in human beings to a significant cause of infection (1). These bacteria are important causes of nosocomial infections, such as urinary tract infections, bacteremia, and endocarditis (2). Recently, enterococci have become considerably resistant to a broad range of antimicrobial agents, particularly glycopeptides, β-lactam, and aminoglycosides (3). Due to inappropriate use of antibiotics in nosocomial infections, the resistance rate is increasing (4). Enterococcus faecalis and E. faecium are two common species isolated from nosocomial infections. The prevalence of antibiotic resistance among E. faecium is higher than in E. faecalis (4). Enterococci are either intrinsically resistant to antibiotics or acquire the resistance genes (5, 6). The resistance is due to inadequate transfer of antibiotics across the cytoplasmic membranes of bacteria. Antibiotics that are effective on the cell wall of the bacteria, such as β-lactam or vancomycin, have a synergistic effect in the treatment of enterococci infections (7). The presence of high-level gentamicin resistant (HLGR) species is globally important due to their multi-resistant nature (8). Unfortunately, enterococci with high-level aminoglycoside-resistance (HLAR) (MIC > 500 µg/mL) do not seem to be sensitive to this synergistic effect, making treatment more difficult (9). HLAR in enterococci is due to aminoglycoside-modifying enzymes (AMEs). The most common genes coding AME are aac(6’)-aph(2”) and aph(3)-IIIa (10). AMEs eliminate the synergism effect of aminoglycosides when combined with a cell-wall-active agent. Aac(6’)aph(2”) is the most common gene causing HLGR in enterococci, and aph(3)-IIIa is common in high-level kanamycin and streptomycin resistance (11). An increase in the prevalence of HLGR has been observed in European, Asian, and South American countries. The prevalence of HLGR isolates recently soared by 10 times, to above 50% in blood culture isolates from E. faecium, which seems to be a rising trend (12).
2. Objectives
Imam Reza hospital in Kermanshah is a referral center in the western part of Iran, with different wards and specialties. The recognition of antimicrobial resistance patterns, particularly in nosocomial infections, can serve as a guideline for selecting suitable treatments and effective antibiotics. The present study was designed to identify the prevalence of, and to compare, the aac(6’)-aph(2”) and aph(3)-IIIa genes and their antimicrobial resistance patterns among E. faecalis and E. faecium isolates from patients at Imam Reza hospital in 2011 - 2012.
3. Patients and Methods
The study was performed on 138 isolates of Enterococcus obtained from patients hospitalized in different wards of Imam Reza hospital during 2011 - 2012. The Enterococcus isolates were identified at the species level based on standard biochemical methods. The antibiotic susceptibility patterns of the isolates were determined by the disk diffusion method according to clinical and laboratory standard institute (CLSI) guidelines (13). The antimicrobial disks used were kanamycin, teicoplanin, streptomycin, imipenem, ciprofloxacin, and ampicillin (MAST, UK). The minimum inhibitory concentrations (MICs) of the aminoglycosides gentamicin, kanamycin, streptomycin, and amikacin were measured by the microbroth dilution method, with antibiotic dilutions ranging from 16 to 8192 µg/mL.
3.1. PCR for Detection of AME Genes
In this study, PCR was employed to detect two AME genes: aac(6’)-aph(2”) and aph(3”)-IIIa, which have been reported to be considerably distributed among enterococci. The DNA of the isolates was extracted using the boiling method (14). PCR reactions were performed in 15 µL volumes with 1.5 mM of MgCl2, 200 µM of each dNTP, 0.5 µM of each primer, 1X PCR buffer, 1 U Taq DNA polymerase, and 100 ng of the enterococcal chromosomal DNA. After heating at 95°C for 5 min, amplification was performed over 30 cycles: 95°C for 30 s, 30 s at the specific annealing temperature for each primer, and 72°C for 30 second, followed by 72°C for 5 minutes (15). The annealing temperature was 50°C for 30 second for the genes. PCR products were analyzed by electrophoresis at 90 V for 60 minutes on 1% agarose gel. Finally, the gel was stained with ethidium bromide, photographed, and analyzed with the gel documentation system (Bio-Rad, Singapore). The sequence of the primers and the size of PCR products are shown in Table 1.
Sequence of Primers and Size of PCR Products
4. Results
Among 138 isolates obtained from patients in different wards, 33 (24.1%) were E. faecium and 63 (46%) were E. faecalis. Enterococcus isolates were identified to the species level based on biochemical tests, and PCR was done in order to investigate HLGR genes.
4.1. Antibiotic Susceptibility Test (Antibiogram)
The antibiotic susceptibility patterns of the isolates were determined by the disk diffusion method according to CLSI guidelines. The rates of enterococcal resistance to antibiotics were: ciprofloxacin 20 (20.8%), teicoplanin 7 (7.3%), imipenem 9 (9.4%), tobramycin 11 (11.4%), kanamycin 77 (80.2%), erythromycin 88 (91.7%), and ampicillin 82 (85.4%). Erythromycin and ampicillin showed the highest level of resistance, while the lowest level was related to teicoplanin.
4.2. Determining MIC50 and MIC90 for Aminoglycosides
Among the aminoglycosides, the highest level of MIC50 was related to kanamycin and gentamicin. For MIC90, kanamycin, gentamicin, and streptomycin were noteworthy. Eighty-nine percent of isolates were diagnosed as HLGR.
4.3. Prevalence of Resistance Genes to Aminoglycosides
The aac(6)-le-aph(2)la and aph(3)-llla genes were determined by PCR according to specific primers. Figure 1 and 2 show the gel electrophoresis results for these genes. Based on the PCR method, the aac(6)-le-aph(2)la and aph(3)-llla genes were found in 64 (61.6%) and 22 (19.6%) of the isolates, respectively. Table 2 shows the frequency of genotypes resistant to aminoglycosides according to enterococcal species.
Gel Electrophoresis of Genus-Specific aac(6)-le-aph(2)la PCR Products From Enterococcal Species
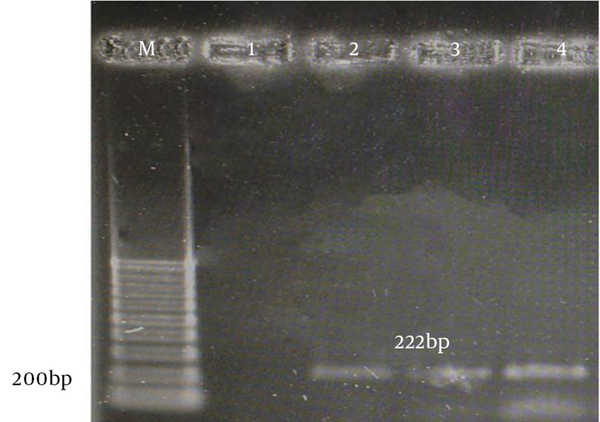
Gel Electrophoresis of Genus-Specific aph(3)-llla PCR Products From Enterococcal Species
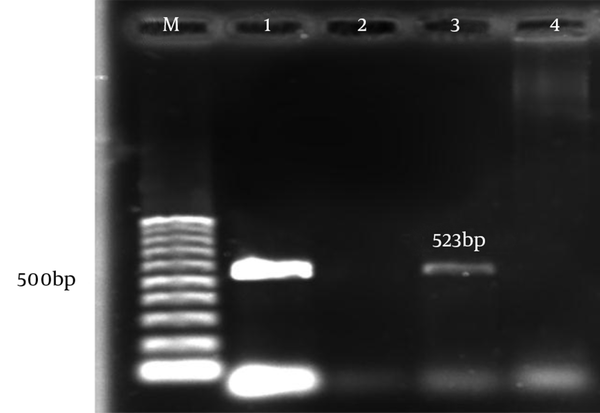
| Gene | E. faecalis | E. faecium |
|---|---|---|
| aac(6)-le-aph(2)la | 43 (67.2) | 21 (32.8) |
| aph(3)-llla | 5 (22.7) | 17 (77.3) |
5. Discussion
In recent years, enterococci have become important causes of nosocomial infections throughout the world, and it has been proven that enterococcal strains are the main cause of 82% of urinary tract infections in Iran (18). Aminoglycosides are frequently used to treat enterococcal infections, which can lead to the spread of aminoglycoside-resistant isolates in Iranian hospitals (19). The increasing number of enterococcal species resistant to antibiotics are currently a challenge with regard to treatment. In the present study, enterococcal isolates obtained from Imam Reza hospital, a referral hospital in Kermanshah, were studied to determine the prevalence of E. faecalis and E. faecium among hospitalized patients. The patterns of antimicrobial resistance of the isolates and two important HLGR genes, aac(6’)-le-aph(2")-la and aph(3), were also investigated. Among the different species of enterococci that are currently recognized, E. faecalis is responsible for 85% - 95% of enterococcal infections, while 5% - 10% of infections are caused by E. faecium (20).
In a similar study in Iran, it has been reported that E. faecalis and E. faecium strains have the highest incidence, 80% - 85% and 15% - 20%, respectively (21, 22). In contrast, European studies have reported that E. faecium was the most prevalent isolate, followed by E. hirae and E. faecalis (23). Aleksandrowicz (24) showed that E. faecalis was the most frequent enterococcal species in all samples, more than 70%, followed by E. faecium (20.4%). In our study, 65.6% of isolates were identified as E. faecalis and 34.4% were E. faecium. Enterococci have become increasingly resistant to a broad range of antimicrobial agents, particularly glycopeptides, ß-lactams, and aminoglycosides (25). Among the resistance mechanisms, HLAR is widely reported, while glycopeptide-resistant or β-lactam-resistant enterococci are prevalent mostly in western countries (26). HLAR leads to the loss of synergy between cell wall synthesis-inhibiting antibiotics, such as penicillins or glycopeptides, which allows aminoglycosides to penetrate through the impermeable bacterial cell wall (27).
In the present study, most isolates were sensitive to imipenem (91.9%), and the rates of resistance to erythromycin, ampicillin, and kanamycin were 63.2%, 58.8%, and 55.1%, respectively. A Turkish study by Akhter et al. (28) showed that imipenem was considered to have the highest level of sensitivity. Compared to our study, Imanifooladi et al. (22) showed that most isolates of enterococci were resistant to erythromycin, tetracycline, and ciprofloxacin. With regard to aminoglycoside resistance, 8.1% of isolates were resistant to tobramycin and 55.1% to kanamycin, while 89% of all isolates were HLGR. In comparison to our study, Emaneini’s survey in Tehran showed 52% of isolates to be HLGR, and Hasani’s 2012 study in Tbariz reported that 60.45% of isolates were HLGR with MICs of ≥ 512 μg/mL (29, 30). Our study showed that HLGR isolates are more popular in Kermanshah. According to the literature, HLGR is more common in Iran, and it is predicted that gentamicin will have no place in the treatment of enterococcal infections. Isolates that express high-level resistance to amikacin, gentamicin, kanamycin, and streptomycin were investigated for the presence of genes encoding the aac(6’)-Ie-aph (2”)Ia and aph(3’)-IIIa enzymes.
The HLAR in enterococci is usually coded by the aac(6’)-aph(2”) gene encoding the aac(6’)-aph(2”) enzyme (31). The PCR results for the isolates demonstrated that 68.3% of E. faecalis and 36.4% of E. faecium isolates contain the aac(6’)-aph(2”) gene. A similar study in Japan showed the prevalence of this gene to be 40.1% among E. faecalis and 12.9% among E. faecium samples (32). In Denmark, 32% of isolates were defined as E. faecalis, and all of them contained this gene (12). According to our study, the prevalence of aph(3) was 23.8% and 21.2% for E. faecalis and E. faecium, respectively. It can be concluded that the most prevalent gene leading to aminoglycoside resistance is aac(6)-aph(2), and in the current study, the presence of this gene was more common than aph(3) in both E. faecalis and E. faecium. The large differences among hospitals in both the use of antimicrobials and the prevalence of resistance indicate the potential for further improvement of antibiotic policies, and possibly for hospital infection-control in order to maintain low resistance levels (1). Physicians are obliged to use antibiotics appropriately and to comply with infection-control policies in an effort to prevent further spread of these resistant organisms.
Acknowledgements
References
-
1.
El-Amin NM, Faidah HS. Vancomycin-resistant Enterococci. Prevalence and risk factors for fecal carriage in patients at tertiary care hospitals. Saudi Med J. 2011;32(9):966-7. [PubMed ID: 21894365].
-
2.
Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun. 2005;73(4):2461-8. [PubMed ID: 15784592]. https://doi.org/10.1128/IAI.73.4.2461-2468.2005.
-
3.
Kristich CJ, Rice LB, Arias CA. Enterococcal Infection-Treatment and Antibiotic Resistance. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston; 2014. [PubMed ID: 24649502].
-
4.
Olawale KO, Fadiora SO, Taiwo SS. Prevalence of hospital-acquired enterococci infections in two primary-care hospitals in osogbo, southwestern Nigeria. Afr J Infect Dis. 2011;5(2):40-6. [PubMed ID: 23878706].
-
5.
Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57(1):138-63. [PubMed ID: 8385262].
-
6.
Simjee S, Gill MJ. Gene transfer, gentamicin resistance and enterococci. J Hosp Infect. 1997;36(4):249-59. [PubMed ID: 9261754].
-
7.
Dupont H, Friggeri A, Touzeau J, Airapetian N, Tinturier F, Lobjoie E, et al. Enterococci increase the morbidity and mortality associated with severe intra-abdominal infections in elderly patients hospitalized in the intensive care unit. J Antimicrob Chemother. 2011;166(10):2379-85. https://doi.org/10.1093/jac/dkr308.
-
8.
Woodford N, Reynolds R, Turton J, Scott F, Sinclair A, Williams A, et al. Two widely disseminated strains of Enterococcus faecalis highly resistant to gentamicin and ciprofloxacin from bacteraemias in the UK and Ireland. J Antimicrob Chemother. 2003;52(4):711-4. [PubMed ID: 12917231]. https://doi.org/10.1093/jac/dkg408.
-
9.
Kafil HS, Asgharzadeh M. Vancomycin-resistant enteroccus faecium and enterococcus faecalis isolated from education hospital of iran. Maedica (Buchar). 2014;9(4):323-7. [PubMed ID: 25705299].
-
10.
Chow JW, Kak V, You I, Kao SJ, Petrin J, Clewell DB, et al. Aminoglycoside resistance genes aph(2")-Ib and aac(6')-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob Agents Chemother. 2001;45(10):2691-4. [PubMed ID: 11557456]. https://doi.org/10.1128/AAC.45.10.2691-2694.2001.
-
11.
Qu TT, Zhang Y, Yu YS, Chen YG, Wei ZQ, Li LJ. [Genotypes of aminoglycoside-modifying enzyme and clinical study of high-level gentamycin resistant enterococcus]. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2006;35(1):76-82. [PubMed ID: 16470925].
-
12.
Rosvoll TC, Lindstad BL, Lunde TM, Hegstad K, Aasnaes B, Hammerum AM, et al. Increased high-level gentamicin resistance in invasive Enterococcus faecium is associated with aac(6')Ie-aph(2'')Ia-encoding transferable megaplasmids hosted by major hospital-adapted lineages. FEMS Immunol Med Microbiol. 2012;66(2):166-76. [PubMed ID: 22672387]. https://doi.org/10.1111/j.1574-695X.2012.00997.x.
-
13.
Hindler JF, Stelling J. Analysis and presentation of cumulative antibiograms: a new consensus guideline from the Clinical and Laboratory Standards Institute. Clin Infect Dis. 2007;44(6):867-73. [PubMed ID: 17304462]. https://doi.org/10.1086/511864.
-
14.
Queipo-Ortuno MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15(2):293-6. [PubMed ID: 18077622]. https://doi.org/10.1128/CVI.00270-07.
-
15.
Rahimi F, Bouzari M, Katouli M, Pourshafie MR. Antibiotic resistance pattern of methicillin resistant and methicillin sensitive Staphylococcus aureus isolates in Tehran, Iran. Jundishapur J Microbio. 2013;6(2):144-9. https://doi.org/10.5812/jjm.4896.
-
16.
Ferretti JJ, Gilmore KS, Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6'-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986;167(2):631-8. [PubMed ID: 3015884].
-
17.
Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos MJ, Lerner SA, Chow JW. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob Agents Chemother. 2003;47(4):1423-6. [PubMed ID: 12654683].
-
18.
Hoseini Zadeh A, Shojapour M, Nazari R, Akbari M, Sofian M, Abtahi H. Genotyping of vancomycin resistant enterococci in arak hospitals. Jundishapur J Microbiol. 2015;8(4). ee16287. [PubMed ID: 26034536]. https://doi.org/10.5812/jjm.8(4)2015.16287.
-
19.
Feizabadi MM, Asadi S, Aliahmadi A, Parvin M, Parastan R, Shayegh M, et al. Drug resistant patterns of enterococci recovered from patients in Tehran during 2000-2003. Int J Antimicrob Agents. 2004;24(5):521-2. [PubMed ID: 15519491]. https://doi.org/10.1016/j.ijantimicag.2004.08.004.
-
20.
Winn W, Allen S, Janda W, Koneman E, Procop G, Schreckenberger PC, et al. Gram-positive cocci part II: streptococci, enterococci, and the “streptococcus-like” bacteria. 6th ed. 2006. p. 673-764.
-
21.
Behnood A, Farajnia S, Moaddab SR, Ahdi-Khosroshahi S, Katayounzadeh A. Prevalence of aac(6')-Ie-aph(2'')-Ia resistance gene and its linkage to Tn5281 in Enterococcus faecalis and Enterococcus faecium isolates from Tabriz hospitals. Iran J Microbiol. 2013;5(3):203-8. [PubMed ID: 24475324].
-
22.
Imanifooladi A, Soltanpour MJ, Kachuei R, Mirnejad R, Rahimi M. The Comparison of Antibacterial Effects of Common Antiseptics against Three Nosocomial Resistant Strains. Med Lab J. 2008;2(1).
-
23.
Borhani K, Ahmadi A, Rahimi F, Pourshafie MR, Talebi M. Determination of Vancomycin Resistant Enterococcus faecium Diversity in Tehran Sewage Using Plasmid Profile, Biochemical Fingerprinting and Antibiotic Resistance. Jundishapur J Microbiol. 2014;7(2). ee8951. [PubMed ID: 25147674]. https://doi.org/10.5812/jjm.8951.
-
24.
Aleksandrowicz I. [The incidence of high-level aminoglicoside and high-level beta-lactam resistance among enterococcal strains of various origin]. Med Dosw Mikrobiol. 2012;64(1):11-8. [PubMed ID: 22808725].
-
25.
Leclercq R. Enterococci acquire new kinds of resistance. Clin Infect Dis. 1997;24 Suppl 1:S80-4. [PubMed ID: 8994783].
-
26.
Stucki K, Harbarth S, Nendaz M. [Enterococcal infections: from simple to most complex]. Rev Med Suisse. 2014;10(446):1918. 1920-3. [PubMed ID: 25438375].
-
27.
Aslangul E, Ruimy R, Chau F, Garry L, Andremont A, Fantin B. Relationship between the level of acquired resistance to gentamicin and synergism with amoxicillin in Enterococcus faecalis. Antimicrob Agents Chemother. 2005;49(10):4144-8. [PubMed ID: 16189091]. https://doi.org/10.1128/AAC.49.10.4144-4148.2005.
-
28.
Akhter S, Asna ZH, Rahman MM. Prevalence and antimicrobial susceptibility of enterococcus species isolated from clinical specimens. Mymensingh Med J. 2011;20(4):694-9. [PubMed ID: 22081191].
-
29.
Emaneini M, Aligholi M, Aminshahi M. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol. 2008;57(2):173-8. [PubMed ID: 18646406].
-
30.
Hasani A, Sharifi Y, Ghotaslou R, Naghili B, Hasani A, Aghazadeh M, et al. Molecular screening of virulence genes in high-level gentamicin-resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens in Northwest Iran. Indian J Med Microbiol. 2012;30(2):175-81. [PubMed ID: 22664433]. https://doi.org/10.4103/0255-0857.96687.
-
31.
Salem-Bekhit MM, Moussa IMI, Muharram MM, Elsherbini AM, AlRejaie S. Increasing prevalence of high-level gentamicin resistant enterococci: An emerging clinical problem. Afr J Microbiol. 2011;5(31):5713-20.
-
32.
Watanabe S, Kobayashi N, Quinones D, Nagashima S, Uehara N, Watanabe N. Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6')-Ie-aph(2'')-Ia or aph(2'')-Ie in a Japanese hospital. Microb Drug Resist. 2009;15(3):185-94. [PubMed ID: 19728776]. https://doi.org/10.1089/mdr.2009.0917.