Published online Apr 26, 2015. doi: 10.4252/wjsc.v7.i3.583
Peer-review started: July 29, 2014
First decision: October 16, 2014
Revised: December 1, 2014
Accepted: December 16, 2014
Article in press: December 17, 2014
Published online: April 26, 2015
Hox genes are an evolutionary highly conserved gene family. They determine the anterior-posterior body axis in bilateral organisms and influence the developmental fate of cells. Embryonic stem cells are usually devoid of any Hox gene expression, but these transcription factors are activated in varying spatial and temporal patterns defining the development of various body regions. In the adult body, Hox genes are among others responsible for driving the differentiation of tissue stem cells towards their respective lineages in order to repair and maintain the correct function of tissues and organs. Due to their involvement in the embryonic and adult body, they have been suggested to be useable for improving stem cell differentiations in vitro and in vivo. In many studies Hox genes have been found as driving factors in stem cell differentiation towards adipogenesis, in lineages involved in bone and joint formation, mainly chondrogenesis and osteogenesis, in cardiovascular lineages including endothelial and smooth muscle cell differentiations, and in neurogenesis. As life expectancy is rising, the demand for tissue reconstruction continues to increase. Stem cells have become an increasingly popular choice for creating therapies in regenerative medicine due to their self-renewal and differentiation potential. Especially mesenchymal stem cells are used more and more frequently due to their easy handling and accessibility, combined with a low tumorgenicity and little ethical concerns. This review therefore intends to summarize to date known correlations between natural Hox gene expression patterns in body tissues and during the differentiation of various stem cells towards their respective lineages with a major focus on mesenchymal stem cell differentiations. This overview shall help to understand the complex interactions of Hox genes and differentiation processes all over the body as well as in vitro for further improvement of stem cell treatments in future regenerative medicine approaches.
Core tip:Hox genes are involved in embryonic development as well as in repair mechanisms in the adult body, thus regulating cell fate. These genes have also been found to be driving factors in various stem cell differentiations in vitro and in vivo which makes them interesting tools for future improvements in stem cell therapies. Therefore, this review outlines the involvement of Hox genes in various stem cell differentiations with a major emphasis on mesenchymal stem cell differentiations.
-
Citation: Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of
Hox genes in stem cell differentiation. World J Stem Cells 2015; 7(3): 583-595 - URL: https://www.wjgnet.com/1948-0210/full/v7/i3/583.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i3.583
The life expectancy of the overall population has increased in most countries, accompanied by an increase in injuries, diseases, and age related degenerative diseases. Thus, reconstructions of hard and soft tissues are an increasing challenge in regenerative medicine[1]. Stem cells could help to overcome these restrictions using their self-renewal and differentiation properties, with or without biomaterials or artificial scaffolds[1-3]. Numerous sources are known from which stem cells can be isolated, often with a varying differentiation potential[4]. Additionally, the commitment might depend on the body site from which the stem cells were isolated[4,5]. In vivo, stem cell fate is determined according to tissues and thus extracellular but also intracellular factors, among them p53, purinergic receptors, collagens, parathyroid hormone-related proteins, or homeobox (Hox) genes[6-9]. The later are the central players in determining regional specificity in the developing embryo and not astonishingly also in numerous differentiations[10].
The Hox gene family consists of 39 members, which are clustered in four paralogous groups and are numbered from 1-13 within the groups. During embryogenesis, Hox genes show an expression pattern which is specific for each body region, commonly referred to as the HOX code. They are an evolutionary highly conserved collection of genes binding to various DNA recognition sites and thus influencing a cell’s developmental fate in a huge variety of species, while determining the anterior-posterior body axis[11] (Figure 1). The precise function of these genes is achieved by their specific temporal and spatial activation over the course of life. Due to this role in embryonic and adult developmental stages, Hox genes are expected to also have fundamental influences on the differentiation of various stem cells in vivo and in vitro towards their respective lineages. There have been studies which directly identified Hox gene involvement in stem cell differentiations, and others which suggest their role based on observed effects of Hox genes in in vivo or in vitro tissue development. It has been speculated that the code may contribute to maintaining the stem cell character in certain niches, since it is also regulated on an epigenetic level[12].
This review will outline the Hox profile of various stem cells, and discuss the wide range of findings concerning Hox genes in stem cell differentiations gathered over the last decade. The nomenclature used for Hox genes in this study is according to the current regulations, meaning that Hox genes related to findings in mice and rats are written with the first letter capitalized, whereas the following are in small capitals (e.g., Hoxc8). For findings in human and chicken, all letters are capitalized (e.g., HOXC8). Understanding the principle behind the role of Hox genes in differentiations and development may provide improved possibilities to alter stem cell fate in vitro and in vivo, and therefore might result in new or improved approaches in regenerative medicine.
Therapeutic approaches are more and more based on the use of mesenchymal stem cells (MSCs), due to their easy accessibility and handling compared to other kinds of stem cells. Hence, we put specific emphasis on the differentiation lineages of MSCs within this review.
Embryonic stem cells (ESCs) can be gained from the inner cell mass of the blastocyst and are therefore derived from an early stage embryo[13,14]. They are characterized by various markers, having the most significant surface markers SSEA-1, SSEA-3, SSEA-4 as well as TRA-1-60 and TRA-1-81[15]. Additionally ESCs are positive for the transcription factors NANOG and OCT3/4[16,17]. Their pluripotency, the potential to differentiate into each cell lineage and all types of cells of the body, and the proliferation rate causes huge interest in this stem cell type[18,19]. An active repression of Hox clusters has been demonstrated in various studies. On the other hand Hox genes are activated when differentiation processes take place[20]. However, it is not known which Hox code exactly commits these cells into the different lineages, yet[20].
Hematopoietic stem cells (HSCs) are the best characterized type of adult stem cells[21]. They can be obtained from bone marrow, cytokine-mobilized peripheral blood, and cord blood[22]. HSCs are characterized with the antigen cell-surface markers CD34, C-KIT, TIE, and CD133/AC133 and they lack CD38, LIN, and CD45RA[23]. These stem cells differentiate into all kinds of blood cell lineages serving the reconstruction of the lympho-hematopoietic system[24]. In general, the HOXA cluster plays a role in hematopoiesis[25], but the expression of HOXB4 which directs transition from the embryonic to the adult hematopoietic program is a major factor as well[21].
Neural crest cells (NCCs) are multipotent cells exclusively present in vertebrates[26]. These cells are not only found in the early embryonic development but are also present in various neural crest-derived tissues in the fetal and adult organism[27]. NCCs are located in between the neural epithelium and epidermis in the neurulating vertebrate embryo[26]. After they have undergone epithelial-mesenchymal transition the NCCs migrate within the embryonic tissue to the respective location and undergo differentiation[28]. This stem cell type commits towards facial cartilage, bone and connective tissue as well as neural and glial cells of the peripheral nervous system. Additionally, they are able to differentiate into skin pigment cells, mesenchyme and in restricted locations into smooth muscle cells[29]. NCCs are characterized with the markers p75NTR, AP-2α and the transcription factors MSX1, MSCX2 and SLUG in the pre-migratory state[30]. In post-migratory state, embryonic and adult cell markers can be distinguished as follows: in the embryo, adult gut and bone marrow p75 is present, whereas SOX10 is found in the dorsal root ganglia of the embryo and in the adult bone marrow. As an additional marker NESTIN is suggested because it is present in the adult heart and embryonic boundary cap cells. Other markers are MUSASHI in the adult heart, SCA1 and CD34 in the adult cornea as well as SLUG and SNAIL in the adult bone marrow[26]. Usually there are no Hox genes expressed in NCCs[31]. However data has been found where Hoxa2 and Hoxd9 can be induced in mouse trunk neural crest cells by retinoic acid, a major factor triggering differentiation[32]. Additionally, Hoxd3 has been shown to have an impact on the formation of somitic mesoderm-and neural crest-derived structures during mouse development[33].
MSCs are multipotent adult stem cells which can be in vitro differentiated towards lineages of mesenchymal tissues[7,34,35], which is in line with their natural repair function in the body where they regenerate mesenchymal tissues like bone, cartilage, muscle ligament, tendon and adipose tissues[7,36]. In addition it has been described that these stem cells can also differentiate towards lineages of the other two germ layers, such as towards endothelial cells, hepatocytes[37], or neurons of the ectoderm[38]. MSCs are found in the stroma of various tissues or organs such as bone marrow, umbilical cord, wisdom teeth and adipose tissues[5,7,39]. Defined characteristics of these stem cells are the plastic adherence under standard culture conditions, the presence of the antigen surface markers CD73, CD90 and 105 and the absence of CD34, CD45, CD14 or CD11b, CD79α or CD19 and HLA-DR[40]. Since Hox genes are involved in organ formation and regeneration, it is not astonishing that the degree of Hox gene expression in various adult stem cell populations differs which enables the discrimination of adult stem cells by the respective Hox expression pattern. Cord blood-derived MSCs (CB-MSCs) and unrestricted somatic stem cells (USSCs) are gained from cord blood. Whereas CB-MSCs are positive for HOXA9, HOXB7, HOXC10 and HOXD6, USSCs resembling the profile of typical ESCs are negative for Hox genes. In CB-MSCs and USSCs it has been demonstrated that this expression code is in line with the differentiation potential of stem cells: CB-MSCs can be differentiated towards the adipogenic lineage whereas USSCs generally can be committed towards all three germ layers and show nevertheless no adipogenic differentiation. HOXB7 and HOXD6 expression has also been shown in bone marrow-derived MSCs that generally have a similar expression profile as MSCs derived from adipose tissue. Amnion- and decidua-derived MSCs can be distinguished using HOXC10 expression[41]. Taken together, a distinct expression of HOXA5 and A10, HOXB6, B7, HOXC4, C6, C8, C9 and C10 as well as HOXD3 and D8 is observed in MSCs derived from different human sources[42]. In murine MSCs the expression of Hoxb2, b5, b7 and Hoxc4 has been shown, which regulate self-renewal and influence the differentiation of other stem cells[43].
Cancer stem cells (CSCs) are cancer cells with the capability of both, self-renewal and differentiation, which are the typical characteristics of any stem cell[44]. These stem cells are multipotent, clonogenic, have the capability to induce new tumors and eventually create heterogeneous cell lineages in tumors[45]. Accordingly they are thought to be responsible for progression of cancer, cancer metastasis, and therapy resistance of cancers[46]. In addition, the recurrence and spreading of cancer might be due to CSCs that remain in residual tumors after surgery and treatment. It was suggested that a tumor contains different stem cell like populations due to reduced cell differentiation within a tumor and this heterogeneous population also includes CSCs[44,45]. DNA damages may render tissue stem cells to become CSCs on their way from a stem cell to a fully differentiated cell. However, instead of dying at the different levels of differentiation like normal tissue stem cells do these cancer precursors do not differentiate normally but accumulate forming tumors[47]. Conclusively, this population differs in marker expression and growth capacities[45]. In general CSCs can be characterized by the presence of the markers CD24, CD29, CD44, CD90, CD133 and CD166[44,45]. In some cancers it has been observed that Hox genes are upregulated which are normally restricted to undifferentiated or proliferative cells, and vice versa a down-regulation has been shown of those Hox genes which are normally present in fully differentiated cells[48]. For instance, HOXA9 seems to play a role in acute myelogenous leukemia[49]. HOXB7, HOXC6 and C9 transcription factors function in intima formation and vascularization in vascular-wall multipotent stem cells, which is in line with vascularization being important for cancer metastasis[50]. Further, colon carcinomas show an overexpression of HOXA4 and HOXD10 which distinguishes colonic stem cells from colon carcinoma cells[51].
Induced pluripotent stem cells (iPS) have similar characteristics as ESCs including pluripotency and immortality[52]. As can be seen in Table 1, both stem cell types are generally negative for Hox gene expression. iPS cells are generated by introducing four defined transcription factors into somatic cells, which are OCT3/4, SOX2, KLF4, c-Myc or NANOG, and Lim2b replacing the later two[53,54]. In the meantime, several somatic cells can be reprogrammed such as fibroblasts, hepatocytes, keratinocytes and circulating T-cells, also with a reduced set of transcription factors, ranging from four factors to one factor with respect of the target cell[55]. Both, ESCs and iPS cells are characterized by the pluripotency markers NANOG, OCT3/4, ERAS, ESG1 and ECAT as well as TRA-1-60[53,56]. As iPS cells can differentiate into the same lineages as ESCs, which is the differentiation into all cell types of the three germ layers[57], the Hox gene expression is similar as well. It is repressed as they are quiescent and up-regulated when the differentiation processes start[58].
| Type of stem cell | Hox expression | Role of Hox |
| HSC | HOXA cluster | Hematopoiesis and its transition from embryonic to adult program |
| HOXB4 | ||
| MSC | HOXA5, 10 | Degree of Hox expression pattern differs in the different sources and determines differentiation potential |
| HOXB6, 7 | ||
| HOXC4, 6, 8, 9, 10 | ||
| HOXD3 and D8 | ||
| ESC | None | Active repression of Hox genes until commitment starts |
| CSC | Various | Overexpression of Hox genes present in undifferentiated cells and down-regulation Hox genes present in differentiated cells |
| HOXA9 | Hematopoiesis | |
| HOXB7, C6, C8 | Vascularization | |
| HOXA4, D10 | Colon carcinoma | |
| NCC | HOXD3 | Formation of somitic mesoderm- and neural crest-derived structures |
| iPS | None | Active repression of Hox genes until commitment starts |
The understanding of adipose tissue as an organ with a detrimental effect on the whole organism, due to its secreted adipokines, led to increased studies during the last decades, since the number of obesity patients rises all over the world. A dysfunction of adipocytes in obese patients can, e.g., lead to diabetes type II and cardiovascular diseases[59], the leading cause of death in industrialized countries. Thus understanding the molecular mechanisms in adipogenesis has therefore gained much importance.
Hox gene expression in developing adipocytes has been studied in the pre-adipogenic committed murine cell line 3T3-L1, where Hoxa4, a7, and Hoxd4 are involved in the differentiation towards adipocytes[60]. In human adipose tissue-derived stem cells (hAT-SC), HOXC8 expression is drastically down-regulated during adipogenesis and almost vanishes in mature adipocytes. However, the down-regulation does not arise from a change of RNA expression, but is rather due to post-translational modifications by miRNAs. Forced expression of HOXC8 in hAT-SC inhibits adipogenesis to a certain extent, indicating that this Hox gene has a functional role during the differentiation of these stem cells towards adipocytes[61].
There is also evidence that HOXA1, A4 and HOXC4 are directly involved in the differentiation and further generation of human white and brown adipose tissue[62]. Additionally, the Hox gene expression pattern is varying in adipocytes derived from different fat deposits within the human body. This pattern is also reflected in the in vitro differentiation of human primary pre-adipocytes isolated from the respective region. For instance pre-adipocytes isolated from the abdominal fat depot show an increased expression of HOXA5 and HOXB8 during differentiation towards adipocytes, as compared to gluteal pre-adipocytes, where HOXA10 expression is higher, and HOXC13 expression is exclusively restricted to gluteal pre-adipocytes[63]. Due to this variation in the expression pattern, a general statement about the Hox gene expression in differentiating adipocytes would not be sensible.
Understanding the principles of Hox signaling in white and brown adipose tissue formation might help prevent health damage in obese patients, such as the development of diabetes and consecutive the metabolic syndrome.
Understanding the development of joints and bones is of interest in many fields of research, including regenerative medicine. However, the complete formation of limbs and other hard tissues involves various types of cells and a complex interplay between them. Three main tissue types can be attributed to bone and joint formation, namely tendons, cartilage and bone. Chondral tissue is the first to develop in limb formation, and its condensation is needed for correct bone development. Of great importance in tissue engineering are the underlying signaling pathways and proteins involved[64]. By investigating the Hox gene expression in chondrocytes and osteoblasts, both, in vitro and in vivo, bone formation and regeneration might be improved in the future, since in both cell types Hoxa2 and Hoxd9 have been shown to be significantly regulated. Hoxa2 is up-regulated in bone regeneration of rats after post-fracture day 10, whereas Hoxd9 is down-regulated after this time period[65]. In mice, Hoxa2 impairs chondrogenesis in the mesenchymal-to-chondrocyte differentiation step leading to a proportionate short stature. Additionally, in a Hoxa2 gain-of-function mice model molecular regulators for an endochondral ossification are decreased[66]. In normal mouse development, Hoxa2 inhibits chondrogenesis and intramembranous ossification as well as bone formation when forcedly expressed in osteoprecursors[67]. Forced Hoxa2 expression in cells committed to undergo chondrogenesis also leads to chondrodysplasia and delayed cartilage mineralization and ossification, but does not affect proliferation of the chondrocytes[68]. Interestingly, Cbfa1 induction might inhibit Hoxa2 caused impairment of osteogenic differentiation in vivo[67]. In addition, overexpression of HOXA2, and expression of HOXA3 and HOXB4, abolishes the ability of neural crest cells in the rostral domain to differentiate into skeletal structures in chicken[69]. The expression of certain Hox genes seems to inhibit skeletogenic differentiation, particularly jaw formation. In cranial neural crest cells there are specific regions where Hox expression or suppression, respectively is essential for appropriate development. In detail Hoxa2 inhibits Sox9, resulting in an impeded development of neural crest cells into ectomesenchymal stem cells and thus no osteochondral progenitors can form in mice. The absence of Hoxa2 leads to a migration of Sox9 into areas where Hoxa2 is normally expressed which induces ectopic chondrogenesis[70]. In addition, Hox genes suppress osteochondrogenesis in more posterior neural crest cells via epigenetic mechanisms[32]. Hoxa2 exerts an inhibitory effect in differentiation of chondrocytes, but is needed during bone formation. This effect is not related to the patterning function of Hoxa2, since it occurs in various tissues independent of their embryonic origin. Moreover, the ossification process itself is not affected by the Hoxa2 misexpression[68].
Hoxc8 as well as Hoxd4 regulate chondrogenesis in a dose dependent manner in mice. Hoxc8 does not block chondrogenesis, but alters the progression of chondrocytes and their differentiation pathway, leading to an accumulation of proliferating precursor cells, which negatively interfere with the progression of differentiation. A transgenic model for Hoxc8 and Hoxd4 signaling in chondrogenesis has been suggested, but the proposed effects have not yet been proven in in vitro culture systems with postnatal derived chondrocytes[71,72].
In Hoxa11 and Hoxd11 double mutant mice, chondrogenesis is initiated normally. Pre-chondrocytes express Sox9 and type II collagen necessary for further differentiation. However, mesenchymal condensations, which later give rise to the radius and ulna, differ in shape and size from non-mutants[73]. Prehypertrophic cells within these condensations do no longer undergo their differentiation pathway towards hypertrophic chondrocytes. Hoxa11 and Hoxd11 double deficient mice have shorter limb formation and development postnatal, together with delayed ossification, suggesting that these two Hox genes are important regulators in mammalian chondrogenesis[73]. This is in line with the finding that HOXA11 is expressed only in tissues which do not undergo chondrogenesis in chicken[74].
Also, Hox genes generate a positional memory in skeletal stem cells, influencing their differentiation potential. Murine skeletal progenitors derived from the mesoderm maintain Hoxa11 expression, whereas mandibular bone progenitors derived from the neural crest are negative for Hox expression. Thus an appropriate expression of Hox genes provides instructions for site-specific differentiation and function of stem cells in vivo and in vitro[75].
Similar as already discussed for the adipogenic lineage commitment, the expression pattern of Hox genes in stem cell differentiations towards the osteogenic lineage varies according to the extraction site of the adipose tissue-derived stem cells. Hoxa4 and a5 expression is decreased in flank-derived MSCs differentiated towards osteoblasts, compared to the level in abdomen-derived stem cells, but Hoxa4 is dominantly expressed in arm-derived stem cells. Hoxc8 and Hoxd4 on the other hand show a similar expression pattern in stem cells derived from the abdomen, arm, flank or thigh[76].
Genes within the D family of the Hox cluster are suggested to play a role in the condensation of MSCs and the further development into chondrocytes. In detail, HOXD9 and HOXD13 were found to be distinctly expressed in developing chondrocytes within MSCs from the chicken limb bud. The inhibition of HOXD10, D11, and D13, respectively, inhibits chondrogenesis[77]. This is in line with the finding that HOXD11 was found to act on initial cartilage condensation phases and later growth phases within the bone of chicken, whereas HOXD13 acts at later stages of bone development. Correspondingly, misexpression of HOXD13 is associated with a shortening of long bones[78]. This is supported by data derived from mice where Hoxd13 and to a lesser extent Hoxd12 were found to be essential for bone formation. A defect in one of these Hox genes leads to limb malformations and shorter bones[79]. Especially Hoxd13 seems to be responsible for the genetic switch from long bone to short bone formation[80]. But also Hoxd10 affects mouse limb development by impairing proper growth of skeletal elements[81]. Large fractures heal by processes very similar to bone formation during development, apart from remodeling and inflammatory responses[5].
The strict Hox gene pattering that could be observed in Drosophila melanogaster is even more complex during bone formation in vertebrae. In D. melanogaster genes closer to the DNA 5’-end are expressed strictly posterior and the more 3’ located Hox genes are strictly anterior. In agreement with that a knockout of Hoxd9 has revealed a more posterior impaired growth in the spine of mice than a knockout of Hoxd13[82].
As already mentioned Hoxd13 is more essential for proper bone formation than Hoxd12, but defects in either of these Hox genes lead to a shortening of bones and limb malformations[79,81]. However, functions missing in knockout mice lacking a certain Hox gene may be compensated by other Hox genes. Hence, the severity when lacking either one of two Hox genes may be far less than if both Hox genes are knocked-out[82,83]. Cases where a Hox gene on both alleles is dysfunctional always show more severe bone malformations[79,81]. Also, stem cells from various sources show an origin-specific Hox expression profile. Therefore, it is of great importance to match the Hox code of the cells subject to transplantation to the tissue of interest. It has been shown that USSCs, which are in vivo Hox negative and lack the potential to differentiate into the adipogenic lineage, gain this potency after these cells have been altered in their Hox expression profile in a co-culture model with CB-MSCs or bone marrow-derived stem cells. This however also affects the chondrogenic and osteogenic differentiation potential of USSCs[84]. The positional memory may hence be altered under the influence of Hox genes, but side effects have to be taken into consideration.
An indirect link between Hox expression and bone formation was examined recently. BMP4 (bone morphogenic protein) as well as BMP7 elevates the expression of KDM6B, which then causes a removal of H3K27me3 that normally inhibits Hox expression. Even though BMP is among other tasks directly responsible for bone formation this secondary function might further aid in the development[85].
A well understood example for how Hox genes are involved in bone formation is HOXA10. Again a BMP, here BMP2 induces HOXA10 which subsequently interacts directly with RUNX2, a main and early regulator of osteogenesis-specific gene expression[86,87]. This is supported by the finding of Gordon and colleagues[88] that Pbx1 is able to repress this Hoxa10-mediated osteogenic differentiation by blocking recruitment of chromatin remodeling factors and that overexpression of Hoxa10 increased the expression of osteoblast-related genes, osteoblast differentiation and mineralization[89].
Once bone and cartilage have been formed, they are connected to the musculature via tendons. Influencing the proper development and regeneration of tendons and striated muscle is therefore of great medical importance. However, little is known to date about the involvement of Hox genes in their developmental processes. The first and only data are about the autopodal Hox genes HOXA13 and HOXD13 inducing Six2 expression, which is a transcription factor present in developing tendon precursors, even in areas where it normally would not occur, such as the zeugopods of a chicken limb-bud[90]. Further investigations are therefore required to be able to improve overall limb regeneration with the help of stem cells under the influence of Hox genes.
Regeneration and repair of the cardiovasculature is a major field of research in the scientific community, since it is the main cause of death with up to 50% in industrialized countries[91]. After transplantation of human bone marrow-derived stem cells into a murine adult heart, a limited number of cells survived and over time showed morphological and molecular changes, resembling the phenotype of host cardiomyocytes[92]. This has been reproduced in various studies and with a variety of donor tissues and hosts in vivo and in vitro[93,94]. Thus MSCs hence provide a valuable source for repairing injured myocardium in theory, but the underlying mechanism of differentiations towards myocytes and the cells of the blood vessels, e.g., endothelial and smooth muscle cells, has to be understood thoroughly to improve the outcome of such treatments. Knowing about changes in Hox gene expression in stem cell types and animal models may in the future help improve the differentiation of MSCs into cardiomyocytes for the transplantation into patients.
HOXA1 deficiency can cause cardiovascular diseases in mouse models as well as in human patients, which suggests its role in cardiomyocte differentiations[75,94-98]. TCDD can inhibit mouse embryonic stem cell differentiation towards cardiomyocytes. During this inhibition Hoxa10, a13, a7, Hoxb9, Hoxc8, c9, Hoxd1, d4, d9, d13 are upregulated, whereas Hoxb1 is down-regulated. In detail, Hoxb4 and Hoxd3 are upregulated with a lower dosage of 10 pmol/L TCDD and down-regulated at a dosage of 100 pmol/L TCDD. Hoxa9, Hoxb3, b8, Hoxc6 are down-regulated at a dosage of 10 pmol/L TCDD whereas upregulated at a dosage of 100 pmol/L TCDD[99]. In accordance to these findings, Hoxa10 has been found to impair murine cardiac differentiation in vivo[100].
In the vasculature, HOXA11 and A13 impact myogenic differentiation, as they repress MyoD expression and therefore initiation of myogenic differentiation in chicken limb muscle precursors transfected with these Hox genes[101]. Retention of Hox gene expression profiles and positional identity has been found in muscle cells and MSCs ex vivo[75,95-98].
The genes of the paralog groups Hox6-10 show a higher expression pattern in smooth muscle cells (SMCs) of the athero-resistant thoracic aorta compared to SMCs of the athero-susceptible aortic arch in mice, rat, and pork. This pattern is consistent in human ESCs which were differentiated into precursors of the thoracic aorta and the aortic arch, respectively, which also suggests a possible involvement of the above mentioned Hox genes in specific SMC differentiation processes of ESCs[102]. In vascular wall-resident multipotent stem cells (VW-MPSCs), HOXB7, HOXC6 and C8 are expressed in high levels compared to human pluripotent ESCs, mature aortic smooth muscle cells and human umbilical cord endothelial cells[50]. Silencing of these Hox genes in VW-MPSCs leads to a reduction in their in-gel (Matrigel) sprouting capacity, hence suggesting a decreased differentiation potential without affecting proliferation[50]. Finally it has been found that in myogenic cells Hox gene expression is down-regulated via hypermethylation[103].
Changes in Hox gene expression over the differentiation time course can also be observed in the stem cell differentiation towards endothelial cells. When mouse ESCs are differentiated towards endothelial cells, Hoxa3 and Hoxd3, which drive angiogenesis and endothelial cell sprouting in adults, show a peak expression at day three of differentiation. Their expression declines over the following days of differentiation, whereas Hoxa5 and Hoxd10 arise from a low expression profile at day three of differentiation to a high expression profile over the course of time. In addition, these two Hox genes are involved in maintaining a mature quiescent endothelial cell phenotype[22]. This suggests an involvement of Hoxa5 and Hoxd10 in the differentiation of ESCs towards endothelial cells, whereas Hoxa3 and Hoxd3 rather mark an immature angiogenic endothelium and/or the onset of angiogenesis[22]. This has been confirmed by others who found that Hoxd10 suppresses angiogenesis, while Hoxd3 and Hoxb3 promote angiogenesis[104,105].
Interestingly, the expression profile of Hox genes in human ESCs from leukemia prone infants is changed which might account for the impairment of hematopoietic cell differentiation in favor of endothelial cell fate. In these cells, HOXA9, A13, HOXB2, B3, B4, B5, and B6 are strongly up-regulated[106]. Next to this, HOXA9 is down-regulated in endothelial cells derived from endothelial progenitor cells from systemic sclerosis patients, suggesting an involvement of this Hox gene in endothelial cell differentiation or mal-differentiation[107].
During the differentiation of human bone marrow-derived MSCs towards endothelial cells four Hox genes were found to be significantly altered in their expression profile. HOXA7 and HOXB3 expression is increased, whereas the expression of HOXA3 and HOXB13 ceased[108]. Also, HOXB5 up-regulates vascular endothelial growth factor receptor-2 which leads to an augmented endothelial cell precursor differentiation. Consequently, HOXB5 has also been proven to enhance endothelial cell sprouting and coordinated vascular growth in chicken[109]. A variety of Hox genes have already been found to influence call fate in favor of endothelial and smooth muscle cell lineages, but the exact mechanisms need to be made clearer in order to use them as transcription factors in regenerative therapies.
Neurodegenerative diseases are also becoming more and more prominent within an aging population. Repairing the neurological systems within the body has been difficult ever since, due to its complexity. A detailed understanding of molecular processes involved in the development and differentiation of neuron precursors is of crucial importance. Since Hox genes control the anterior-posterior (AP) segmentation, the 3’ genes in the Hox cluster are more prone to play a role in neurogenesis, and the paralog groups 1-4 have restricted expression borders in the hindbrain. The remaining paralog groups 5-13 reveal an anterior expression border in the spinal cord. This AP segmentation translates to the migration of neural crest cells[12].
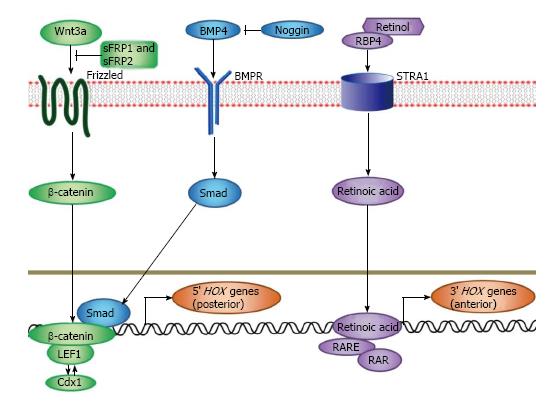
In mouse models, Hoxb1 is essential for the correct development of rhombomeres at early embryonic stages, since it naturally induces neural stem cells towards a hindbrain identity, and has also been shown to be involved in the differentiation, maturation and maintenance of facial nerves[110,111]. Murine Hoxa2 and Hoxb1 can individually and as the sole additional factor induce motor neuron development, even in sections where they are usually not present[112]. In mice deficient for Hoxb1 expression, facial neurons do not properly form[113]. Also, HOXB4 has been found to be a crucial factor in driving neuronal differentiations in the neural tube[114]. Further, since the disruption of Hoxa1 in mice leads to malformations of the hindbrain, a role for this Hox gene in murine neurogenesis has been suggested[115,116]. Retinoic acid (RA) has been established as an essential factor for adult neurogenesis. Embryonic stem cells treated with RA almost exclusively differentiate into neurons and develop a Hox expression profile[117]. Many Hox genes contain retinoic acid response elements (RAREs) which supports the finding that they are involved in RA-induced differentiation of ESCs[118]. RAREs have been found in Hoxa1, a4, Hoxb1, b4, and Hoxd4[12,119-128]. In fact, several Hox genes have been found to be strongly up-regulated in differentiations where RA was present[118]. The interaction between RA, Hox genes, and other signaling molecules has been visualized in Figure 2. During embryonic stem cell differentiation, highly conserved elements in the Hox gene clusters are epigenetically modified, so that adult spinal cord progenitors express a distinct Hox expression profile[129-131]. The further investigation of Hox genes in neurogenesis therefore promises a better insight into the possible therapies to treat neurodegenerative diseases and should be pursued.
Degenerative diseases are one of the major problems arising within a population with high average age. The complex biological systems in the human body have made the development of regenerative therapies a challenging project. Much effort has been put into using stem cells and various signaling molecules for this purpose over the last decades. In this review, we have discussed the expression of Hox transcription factors in various types of stem cells and their role in differentiation. Interestingly already the stem cell source has an impact on the Hox gene expression in the respective stem cell type. Hox genes provide valuable possibilities to influence stem cell differentiations. Either single Hox genes or small groups have been found to directly influence the course of several differentiations, but detailed molecular changes under this influence are mostly not yet known. Additionally, many Hox genes have only revealed an indirect effect, or have been investigated in in vivo development but not in stem cell differentiations. Future studies need to be performed to investigate whether the suggested Hox genes, as summarized in Table 2 for mesenchymal stem cell lineages, actually have a functional role in the in vitro differentiation of stem cells. Additionally, the effects must be tested in vivo to evaluate their therapeutic potential in tissue repair and regeneration[132]. As a consequence of the observed results it is suggested that cells which have their HOX code deleted regain plasticity and can be reprogrammed, similar as it has already been shown in iPS cells[133,134]. In addition to this and similar to iPS cells, also the epigenetic profile must be considered. We summarized that next to the histological compatibility of donor and host tissues the HOX code must be taken into consideration in regenerative therapies. In conclusion, the further investigation of Hox genes in stem cell differentiations is an interesting research field in regenerative medicine to improve tissue repair in injury, disease and aging.
| Differentiation | Hox gene expression | Ref. |
| Adipogenesis | a4, a7, d4, A1, A4, A5, A10, B8, C4, C8 ↓, C13 | [60-63] |
| Chondrogenesis | a2 ↓, a11, c8 ↓, d4 ↓, d11, D9 ↑, D11, D13 ↑ | [65,66,70-73,77,78] |
| Osteogenesis | a2 ↑, a10, d9 ↓, d12, d13, A10 | [82,86,87,89] |
| Tendon differentiation | A13, D13 | [90] |
| Cardiomyocyte differentiation | a10, A1 | [75,95-98,100] |
| Smooth muscle cell differentiation | a6-10, b6-10, c6-10, d6-10, A11, A13, B7, C6, C8 | [50,101,102] |
| Endothelial cell differentiation | a3 ↓, a5 ↑, d3 ↓, d10 ↑, A7 ↑, A9 ↑, A13 ↑, B2 ↑, B3 ↑, B4 ↑, B5 ↑, B6 ↑, B13 ↑ | [20,106-108] |
| Neurogenesis | a2, b1, B4 | [110-112,114-116] |
P- Reviewer: Rall K S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Zippel N, Schulze M, Tobiasch E. Biomaterials and mesenchymal stem cells for regenerative medicine. Recent Pat Biotechnol. 2010;4:1-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 2. | Schulze M, Tobiasch E. Artificial scaffolds and mesenchymal stem cells for hard tissues. Adv Biochem Eng Biotechnol. 2012;126:153-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Babczyk P, Conzendorf C, Klose J, Schulze M, Harre K, Tobiasch E. Stem Cells on Biomaterials for Synthetic Grafts to Promote Vascular Healing. J Clin Med. 2014;3:39-87. [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 4. | Tobiasch E. Adult Human Mesenchymal Stem Cells as Source for Future Tissue Engineering. 1st ed. Forschungsspitzen und Spitzenforschung. Heidelberg: Physica-Verlag HD 2008; 329-338. [DOI] [Cited in This Article: ] |
| 5. | Khan D, Kleinfeld C, Winter M, Tobiasch E. Oral Tissues as Source for Bone Regeneration in Dental Implantology. 1st ed. Tissue Regeneration. From Basic Biology to Clinical Application. Mexico City: InTech 2012; 325-338. [DOI] [Cited in This Article: ] |
| 6. | Zhang Y, Tobiasch E. The role of purinergic receptors in stem cells and their consecutive tissues. 1st ed. Adult progenitor cell standardization. Aalborg: River Publishers 2011; 73-98. [Cited in This Article: ] |
| 7. | Zippel N, Limbach CA, Ratajski N, Urban C, Luparello C, Pansky A, Kassack MU, Tobiasch E. Purinergic receptors influence the differentiation of human mesenchymal stem cells. Stem Cells Dev. 2012;21:884-900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Longo A, Tobiasch E, Luparello C. Type V collagen counteracts osteo-differentiation of human mesenchymal stem cells. Biologicals. 2014;42:294-297. [PubMed] [Cited in This Article: ] |
| 9. | Zhang Y, Lau P, Pansky A, Kassack M, Hemmersbach R, Tobiasch E. The influence of simulated microgravity on purinergic signaling is different between individual culture and endothelial and smooth muscle cell coculture. Biomed Res Int. 2014;2014:413708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Myers P. Hox genes in development: The Hox code. Nature Education. 2008;1:2. [Cited in This Article: ] |
| 11. | Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 605] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 12. | Barber BA, Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat. 2010;192:261-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5956] [Cited by in F6Publishing: 5269] [Article Influence: 122.5] [Reference Citation Analysis (0)] |
| 14. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11399] [Cited by in F6Publishing: 10061] [Article Influence: 387.0] [Reference Citation Analysis (0)] |
| 15. | Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci USA. 1998;95:13726-13731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1002] [Cited by in F6Publishing: 842] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 16. | Schöler HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543-2550. [PubMed] [Cited in This Article: ] |
| 17. | Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643-655. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2459] [Cited by in F6Publishing: 2344] [Article Influence: 111.6] [Reference Citation Analysis (0)] |
| 18. | Smith AG. Embryonic Stem Cells. Stem Cell Biology. New York: Cold Spring Harbor Laboratory Press 2001; 205-230. [Cited in This Article: ] |
| 19. | Melton DA, Cowan C. Stemness: Definitions, Criteria, and Standards. Essentials of Stem Biology. 2nd ed. San Diego: Academic Press 2009; 22-24. [DOI] [Cited in This Article: ] |
| 20. | Bahrami SB, Veiseh M, Dunn AA, Boudreau NJ. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell Adh Migr. 2011;5:133-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Daley GW. Hematopoetic Stem Cells. Essentials of Stem Cell Biology. 2nd ed. Massachusetts: Academic Press 2009; 211-217. [DOI] [Cited in This Article: ] |
| 22. | André-Schmutz I, Cavazzana-Calvo . Gene and Cell Therapy Involving Hematopoetic Stem Cell. Hematopoietic Stem Cell Development. New York: Kluwer Academic/Plenum Publishers 2006; 154-173. [DOI] [Cited in This Article: ] |
| 23. | Yokota T, Oritani K, Butz S, Ewers S, Vestweber D, Kanakura Y. Markers for Hematopoietic Stem Cells: Histories and Recent Achievements. Advances in Hematopoietic Stem Cell Research. Mexico City: InTech 2012; 77-88. [DOI] [Cited in This Article: ] |
| 24. | Murphy K. Basic Concepts in Immunology. Janeway’s Immunobiology. 8th ed. New York: Garland Science, Taylor & Francis Group 2012; 1-36. [Cited in This Article: ] |
| 25. | Mulgrew NM, Kettyle LM, Ramsey JM, Cull S, Smyth LJ, Mervyn DM, Bijl JJ, Thompson A. c-Met inhibition in a HOXA9/Meis1 model of CN-AML. Dev Dyn. 2014;243:172-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Dupin E, Sommer L. Neural crest progenitors and stem cells: from early development to adulthood. Dev Biol. 2012;366:83-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 27. | Shakhova O, Sommer L. Neural crest-derived stem cells. StemBook. 2010;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Dupin E, Creuzet S, Le Douarin NM. The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol. 2006;589:96-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 84] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Gajavelli S, Wood PM, Pennica D, Whittemore SR, Tsoulfas P. BMP signaling initiates a neural crest differentiation program in embryonic rat CNS stem cells. Exp Neurol. 2004;188:205-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Schwarz D, Varum S, Zemke M, Schöler A, Baggiolini A, Draganova K, Koseki H, Schübeler D, Sommer L. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141:867-877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 32. | Ishikawa S, Ito K. Plasticity and regulatory mechanisms of Hox gene expression in mouse neural crest cells. Cell Tissue Res. 2009;337:381-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Taniguchi Y, Tanaka O, Sekiguchi M, Takekoshi S, Tsukamoto H, Kimura M, Imai K, Inoko H. Enforced expression of the transcription factor HOXD3 under the control of the Wnt1 regulatory element modulates cell adhesion properties in the developing mouse neural tube. J Anat. 2011;219:589-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 34. | Pansky A, Roitzheim B, Tobiasch E. Differentiation potential of adult human mesenchymal stem cells. Clin Lab. 2007;53:81-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Tobiasch E. Differentiation potential of adult human mesenchymal stem cells. Stem Cell Engineering. Heidelberg: Springer 2010; 61-77. [Cited in This Article: ] |
| 36. | Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739-2749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1697] [Cited by in F6Publishing: 1616] [Article Influence: 95.1] [Reference Citation Analysis (0)] |
| 37. | Snykers S, De Kock J, Tamara V, Rogiers V. Hepatic differentiation of mesenchymal stem cells: in vitro strategies. Methods Mol Biol. 2011;698:305-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Zeng R, Wang LW, Hu ZB, Guo WT, Wei JS, Lin H, Sun X, Chen LX, Yang LJ. Differentiation of human bone marrow mesenchymal stem cells into neuron-like cells in vitro. Spine (Phila Pa 1976). 2011;36:997-1005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 39. | Haddouti EM, Skroch M, Zippel N, Müller C, Birova B, Pansky A, Kleinfeld C, Winter M, Tobiasch E. Human dental follicle precursor cells of wisdom teeth: Isolation and differentiation towards osteoblasts for implants with and without scaffolds. Mat Sci Engineer Technol. 2009;40:732-737. [DOI] [Cited in This Article: ] |
| 40. | Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11055] [Cited by in F6Publishing: 11772] [Article Influence: 692.5] [Reference Citation Analysis (1)] |
| 41. | Liedtke S, Buchheiser A, Bosch J, Bosse F, Kruse F, Zhao X, Santourlidis S, Kögler G. The HOX Code as a “biological fingerprint” to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 2010;5:40-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 42. | Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol Cell Biol. 2013;33:3274-3285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Phinney DG, Gray AJ, Hill K, Pandey A. Murine mesenchymal and embryonic stem cells express a similar Hox gene profile. Biochem Biophys Res Commun. 2005;338:1759-1765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 44. | Wang P, Gao Q, Suo Z, Munthe E, Solberg S, Ma L, Wang M, Westerdaal NA, Kvalheim G, Gaudernack G. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS One. 2013;8:e57020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 45. | Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427-13432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 528] [Cited by in F6Publishing: 565] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 46. | Williams K, Giridhar PV, Kasper S. CD44 Integrated Signaling in Stem Cell Microenvironments. Role of Cancer Stem Cells in Cancer Biology and Therapy. Florida: CRC Press 2013; 129-150. [DOI] [Cited in This Article: ] |
| 47. | Sell S. On the stem cell origin of cancer. Am J Pathol. 2010;176:2584-2494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 48. | Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777-785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 436] [Cited by in F6Publishing: 458] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 49. | Velu CS, Chaubey A, Phelan JD, Horman SR, Wunderlich M, Guzman ML, Jegga AG, Zeleznik-Le NJ, Chen J, Mulloy JC. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J Clin Invest. 2014;124:222-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 50. | Klein D, Benchellal M, Kleff V, Jakob HG, Ergün S. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci Rep. 2013;3:2178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Bhatlekar S, Addya S, Salunek M, Orr CR, Surrey S, McKenzie S, Fields JZ, Boman BM. Identification of a developmental gene expression signature, including HOX genes, for the normal human colonic crypt stem cell niche: overexpression of the signature parallels stem cell overpopulation during colon tumorigenesis. Stem Cells Dev. 2014;23:167-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 52. | Tanabe K, Takahashi K, Yamanaka S. Induction of pluripotency by defined factors. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:83-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 53. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17040] [Article Influence: 946.7] [Reference Citation Analysis (0)] |
| 54. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7589] [Cited by in F6Publishing: 6999] [Article Influence: 411.7] [Reference Citation Analysis (0)] |
| 55. | de Lázaro I, Yilmazer A, Kostarelos K. Induced pluripotent stem (iPS) cells: a new source for cell-based therapeutics? J Control Release. 2014;185:37-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 56. | Shimamoto R, Amano N, Ichisaka T, Watanabe A, Yamanaka S, Okita K. Generation and characterization of induced pluripotent stem cells from Aid-deficient mice. PLoS One. 2014;9:e94735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 57. | Qin J, Sontag S, Lin Q, Mitzka S, Leisten I, Schneider RK, Wang X, Jauch A, Peitz M, Brüstle O. Cell fusion enhances mesendodermal differentiation of human induced pluripotent stem cells. Stem Cells Dev. 2014;23:2875-2882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 58. | Atkinson SP, Koch CM, Clelland GK, Willcox S, Fowler JC, Stewart R, Lako M, Dunham I, Armstrong L. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008;26:1174-1185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Hajer GR, van Haeften TW, Visseren FL. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur Heart J. 2008;29:2959-2971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 919] [Cited by in F6Publishing: 943] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 60. | Cowherd RM, Lyle RE, Miller CP, Mcgehee RE. Developmental profile of homeobox gene expression during 3T3-L1 adipogenesis. Biochem Biophys Res Commun. 1997;237:470-475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 61. | Mori M, Nakagami H, Kaneda Y. Homeobox C8 suppresses adipogenesis of human adipose tissue-derived stem cells. Circulation. 2008;118:279. [Cited in This Article: ] |
| 62. | Cantile M, Procino A, D’Armiento M, Cindolo L, Cillo C. HOX gene network is involved in the transcriptional regulation of in vivo human adipogenesis. J Cell Physiol. 2003;194:225-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 63. | Karastergiou K, Fried SK, Xie H, Lee MJ, Divoux A, Rosencrantz MA, Chang RJ, Smith SR. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J Clin Endocrinol Metab. 2013;98:362-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 64. | Grotheer V, Schulze M, Tobiasch E. Trends in Bone Tissue Engineering: Proteins for Osteogenic Differentiation and the Respective Scaffolding. Protein Purification - Principles and Trends. Hong Kong: iConcept Press Ltd 2014; . [Cited in This Article: ] |
| 65. | Gersch RP, Lombardo F, McGovern SC, Hadjiargyrou M. Reactivation of Hox gene expression during bone regeneration. J Orthop Res. 2005;23:882-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Deprez PM, Nichane MG, Lengelé BG, Rezsöhazy R, Nyssen-Behets C. Molecular study of a Hoxa2 gain-of-function in chondrogenesis: a model of idiopathic proportionate short stature. Int J Mol Sci. 2013;14:20386-20398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 67. | Kanzler B, Kuschert SJ, Liu YH, Mallo M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development. 1998;125:2587-2597. [PubMed] [Cited in This Article: ] |
| 68. | Massip L, Ectors F, Deprez P, Maleki M, Behets C, Lengelé B, Delahaut P, Picard J, Rezsöhazy R. Expression of Hoxa2 in cells entering chondrogenesis impairs overall cartilage development. Differentiation. 2007;75:256-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 69. | Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301-4313. [PubMed] [Cited in This Article: ] |
| 70. | Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013;5:a008326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 71. | Yueh YG, Gardner DP, Kappen C. Evidence for regulation of cartilage differentiation by the homeobox gene Hoxc-8. Proc Natl Acad Sci USA. 1998;95:9956-9961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 72. | Cormier SA, Mello MA, Kappen C. Normal proliferation and differentiation of Hoxc-8 transgenic chondrocytes in vitro. BMC Dev Biol. 2003;3:4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 73. | Boulet AM, Capecchi MR. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development. 2004;131:299-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 74. | Rogina B, Coelho CN, Kosher RA, Upholt WB. The pattern of expression of the chicken homolog of HOX1I in the developing limb suggests a possible role in the ectodermal inhibition of chondrogenesis. Dev Dyn. 1992;193:92-101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 75. | Cerdá-Esteban N, Spagnoli FM. Glimpse into Hox and tale regulation of cell differentiation and reprogramming. Dev Dyn. 2014;243:76-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 76. | Levi B, James AW, Glotzbach JP, Wan DC, Commons GW, Longaker MT. Depot-specific variation in the osteogenic and adipogenic potential of human adipose-derived stromal cells. Plast Reconstr Surg. 2010;126:822-834. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 77. | Jung JC, Tsonis PA. Role of 5’ HoxD genes in chondrogenesis in vitro. Int J Dev Biol. 1998;42:609-615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 78. | Goff DJ, Tabin CJ. Analysis of Hoxd-13 and Hoxd-11 misexpression in chick limb buds reveals that Hox genes affect both bone condensation and growth. Development. 1997;124:627-636. [PubMed] [Cited in This Article: ] |
| 79. | Davis AP, Capecchi MR. A mutational analysis of the 5’ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175-1185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 80. | Yokouchi Y, Nakazato S, Yamamoto M, Goto Y, Kameda T, Iba H, Kuroiwa A. Misexpression of Hoxa-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes Dev. 1995;9:2509-2522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 177] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 81. | Carpenter EM, Goddard JM, Davis AP, Nguyen TP, Capecchi MR. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505-4514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 89] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 82. | Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Doll e P, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996;122:461-472. [PubMed] [Cited in This Article: ] |
| 83. | Favier B, Dollé P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 149] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 84. | Liedtke S, Freytag EM, Bosch J, Houben AP, Radke TF, Deenen R, Köhrer K, Kögler G. Neonatal mesenchymal-like cells adapt to surrounding cells. Stem Cell Res. 2013;11:634-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 85. | Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH, Wang CY. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 86. | Balint E, Lapointe D, Drissi H, van der Meijden C, Young DW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem. 2003;89:401-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 126] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 87. | Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337-3352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 130] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 88. | Gordon JA, Hassan MQ, Saini S, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol. 2010;30:3531-3541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 89. | Gordon JA, Hassan MQ, Koss M, Montecino M, Selleri L, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs. 2011;194:146-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 90. | Yamamoto-Shiraishi Y, Kuroiwa A. Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev Biol. 2013;377:363-374. [PubMed] [Cited in This Article: ] |
| 91. | Nature Publishing Group. Cardiovascular disease. Nature Biotechnology Oct. 2000;. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 92. | Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1647] [Cited by in F6Publishing: 1524] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 93. | Xu W, Zhang X, Qian H, Zhu W, Sun X, Hu J, Zhou H, Chen Y. Mesenchymal stem cells from adult human bone marrow differentiate into a cardiomyocyte phenotype in vitro. Exp Biol Med (Maywood). 2004;229:623-631. [PubMed] [Cited in This Article: ] |
| 94. | Potdar PD, Prasannan P. Differentiation of human dermal mesenchymal stem cells into cardiomyocytes by treatment with 5-azacytidine: Concept for regenerative therapy in myocardial infarction. ISRN Stem Cells. 2013;2013:9. [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 95. | Donoghue MJ, Morris-Valero R, Johnson YR, Merlie JP, Sanes JR. Mammalian muscle cells bear a cell-autonomous, heritable memory of their rostrocaudal position. Cell. 1992;69:67-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 96. | Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2005;37:1035-1037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 216] [Cited by in F6Publishing: 198] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 97. | Chi JT, Rodriguez EH, Wang Z, Nuyten DS, Mukherjee S, van de Rijn M, van de Vijver MJ, Hastie T, Brown PO. Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers. PLoS Genet. 2007;3:1770-1784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 98. | Makki N, Capecchi MR. Cardiovascular defects in a mouse model of HOXA1 syndrome. Hum Mol Genet. 2012;21:26-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 99. | Wang Y, Fan Y, Puga A. Dioxin exposure disrupts the differentiation of mouse embryonic stem cells into cardiomyocytes. Toxicol Sci. 2010;115:225-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 100. | Behrens AN, Iacovino M, Lohr JL, Ren Y, Zierold C, Harvey RP, Kyba M, Garry DJ, Martin CM. Nkx2-5 mediates differential cardiac differentiation through interaction with Hoxa10. Stem Cells Dev. 2013;22:2211-2220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 101. | Yamamoto M, Kuroiwa A. Hoxa-11 and Hoxa-13 are involved in repression of MyoD during limb muscle development. Dev Growth Differ. 2003;45:485-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 102. | Trigueros-Motos L, González-Granado JM, Cheung C, Fernández P, Sánchez-Cabo F, Dopazo A, Sinha S, Andrés V. Embryological-origin-dependent differences in homeobox expression in adult aorta: role in regional phenotypic variability and regulation of NF-κB activity. Arterioscler Thromb Vasc Biol. 2013;33:1248-1256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 103. | Tsumagari K, Baribault C, Terragni J, Chandra S, Renshaw C, Sun Z, Song L, Crawford GE, Pradhan S, Lacey M. DNA methylation and differentiation: HOX genes in muscle cells. Epigenetics Chromatin. 2013;6:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 104. | Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N. Sustained expression of homeobox D10 inhibits angiogenesis. Am J Pathol. 2002;161:2099-2109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 113] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 105. | Plummer PN, Freeman R, Taft RJ, Vider J, Sax M, Umer BA, Gao D, Johns C, Mattick JS, Wilton SD. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013;73:341-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 655] [Reference Citation Analysis (0)] |
| 106. | Bueno C, Montes R, Melen GJ, Ramos-Mejia V, Real PJ, Ayllón V, Sanchez L, Ligero G, Gutierrez-Aranda I, Fernández AF. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res. 2012;22:986-1002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 107. | Avouac J, Cagnard N, Distler JH, Schoindre Y, Ruiz B, Couraud PO, Uzan G, Boileau C, Chiocchia G, Allanore Y. Insights into the pathogenesis of systemic sclerosis based on the gene expression profile of progenitor-derived endothelial cells. Arthritis Rheum. 2011;63:3552-3562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 108. | Chung N, Jee BK, Chae SW, Jeon YW, Lee KH, Rha HK. HOX gene analysis of endothelial cell differentiation in human bone marrow-derived mesenchymal stem cells. Mol Biol Rep. 2009;36:227-235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 109. | Winnik S, Klinkert M, Kurz H, Zoeller C, Heinke J, Wu Y, Bode C, Patterson C, Moser M. HoxB5 induces endothelial sprouting in vitro and modifies intussusceptive angiogenesis in vivo involving angiopoietin-2. Cardiovasc Res. 2009;83:558-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 110. | Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630-634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 333] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 111. | Briscoe J, Wilkinson DG. Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 2004;18:1643-1648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 112. | Jungbluth S, Bell E, Lumsden A. Specification of distinct motor neuron identities by the singular activities of individual Hox genes. Development. 1999;126:2751-2758. [PubMed] [Cited in This Article: ] |
| 113. | Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027-5040. [PubMed] [Cited in This Article: ] |
| 114. | Akin ZN, Nazarali AJ. Hox genes and their candidate downstream targets in the developing central nervous system. Cell Mol Neurobiol. 2005;25:697-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 115. | Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR. Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development. 1993;118:1063-1075. [PubMed] [Cited in This Article: ] |
| 116. | Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105-1119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 529] [Cited by in F6Publishing: 565] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 117. | Loring JF, Porter JG, Seilhammer J, Kaser MR, Wesselschmidt R. A gene expression profile of embryonic stem cells and embryonic stem cell-derived neurons. Restor Neurol Neurosci. 2001;18:81-88. [PubMed] [Cited in This Article: ] |
| 118. | Shahhoseini M, Taghizadeh Z, Hatami M, Baharvand H. Retinoic acid dependent histone 3 demethylation of the clustered HOX genes during neural differentiation of human embryonic stem cells. Biochem Cell Biol. 2013;91:116-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 119. | Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3’ of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38:217-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 120. | Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 370] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 121. | Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728-1732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 235] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 122. | Dupé V, Davenne M, Brocard J, Dollé P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3’ retinoic acid response element (3’RARE). Development. 1997;124:399-410. [PubMed] [Cited in This Article: ] |
| 123. | Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 242] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 124. | Packer AI, Crotty DA, Elwell VA, Wolgemuth DJ. Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development. 1998;125:1991-1998. [PubMed] [Cited in This Article: ] |
| 125. | Zhang F, Nagy Kovács E, Featherstone MS. Murine hoxd4 expression in the CNS requires multiple elements including a retinoic acid response element. Mech Dev. 2000;96:79-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 126. | Huang D, Chen SW, Gudas LJ. Analysis of two distinct retinoic acid response elements in the homeobox gene Hoxb1 in transgenic mice. Dev Dyn. 2002;223:353-370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 127. | Rastegar M, Kobrossy L, Kovacs EN, Rambaldi I, Featherstone M. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol Cell Biol. 2004;24:8090-8103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 128. | Nolte C, Rastegar M, Amores A, Bouchard M, Grote D, Maas R, Kovacs EN, Postlethwait J, Rambaldi I, Rowan S. Stereospecificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Dev Biol. 2006;299:582-593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 129. | Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119-1130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 491] [Cited by in F6Publishing: 491] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 130. | Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3971] [Cited by in F6Publishing: 3874] [Article Influence: 215.2] [Reference Citation Analysis (0)] |
| 131. | Leung C, Chan SC, Tsang SL, Wu W, Sham MH. Cyp26b1 mediates differential neurogenicity in axial-specific populations of adult spinal cord progenitor cells. Stem Cells Dev. 2012;21:2252-2261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 132. | Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 133. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3332] [Cited by in F6Publishing: 3002] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 134. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2048] [Cited by in F6Publishing: 1876] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 135. | Abdel-Fattah R, Xiao A, Bomgardner D, Pease CS, Lopes MB, Hussaini IM. Differential expression of HOX genes in neoplastic and non-neoplastic human astrocytes. J Pathol. 2006;209:15-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 136. | Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol. 2006;16:2206-2216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 137. | Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, Joseph JD, McDonnell DP. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 138. | Chuai M, Weijer CJ. Regulation of cell migration during chick gastrulation. Curr Opin Genet Dev. 2009;19:343-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 139. | Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965-979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 140. | Falaschi A, Abdurashidova G, Biamonti G. DNA replication, development and cancer: a homeotic connection? Crit Rev Biochem Mol Biol. 2010;45:14-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 141. | Béland M, Pilon N, Houle M, Oh K, Sylvestre JR, Prinos P, Lohnes D. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol Cell Biol. 2004;24:5028-5038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 142. | DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 2000;8:309-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 616] [Cited by in F6Publishing: 560] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 143. | Gazit A, Yaniv A, Bafico A, Pramila T, Igarashi M, Kitajewski J, Aaronson SA. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959-5966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 144. | Gouti M, Gavalas A. Hoxb1 controls cell fate specification and proliferative capacity of neural stem and progenitor cells. Stem Cells. 2008;26:1985-1997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 145. | Kim JH, Liu X, Wang J, Chen X, Zhang H, Kim SH, Cui J, Li R, Zhang W, Kong Y. Wnt signaling in bone formation and its therapeutic potential for bone diseases. Ther Adv Musculoskelet Dis. 2013;5:13-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 234] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 146. | Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JB. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 147. | Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 500] [Cited by in F6Publishing: 503] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 148. | Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816-818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 286] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 149. | Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 150. | Wawrzak D, Métioui M, Willems E, Hendrickx M, de Genst E, Leyns L. Wnt3a binds to several sFRPs in the nanomolar range. Biochem Biophys Res Commun. 2007;357:1119-1123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |