Real-time 3D image reconstruction guidance in liver resection surgery
Introduction
Surgery today has the best survival rate of patients suffering from hepatic cancer. However, eligibility for hepatic surgery, which relies on various criteria and rules for partial resection and transplant, limits to less than 50% patients who can be operated. Accurate knowledge of liver anatomy is thus a key point for any surgical procedure, including hepatic tumor resection or transplant from living donor. Nowadays, fundamental anatomical descriptions by Couinaud (1) and Bismuth (2) are largely accepted to describe the relevant segmental hepatic anatomy in surgery. This basic anatomy can be identified on medical imaging, usually computed tomography (CT) or MRI images. Although these images contain all required information in tumors, major vessels and biliary tracts, surgeons can find it difficult to perceive relations of these structures before surgery, during surgical planning. It seems thus fundamental to offer surgeons tools that will ease the interpretation of conventional images. Among these tools, 3D visualization showed significant advantages compared to standard visualization of 2D slices (3,4).
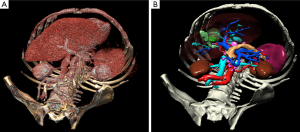
The most common way to analyze medical images in 3D consists in visualizing the data with a direct volume rendering technique (Figure 1). This technique is widely available on workstations in radiology departments and can be accessed via free software on the Internet, such as OsiriX (5) (limited to MacOS) or VR-Render (6) ©IRCAD2008 (running under Windows, MacOS and Linux). This technique can be sufficient to provide good 3D visualization of anatomical and pathological structures. Volume of organs and pathologies is however not available, and volume rendering makes it difficult to virtually resect an organ without entirely cutting neighboring structures. In order to overcome this limit, each anatomical and pathological structure of the medical image has to be recognized and delineated. Resulting 3D models can then be visualized individually thanks to surface rendering (Figure 1). This second solution is more adapted to surgical use, preoperatively for planning and intraoperatively for guidance, each structure having then a different color.
There is today a large number of software tools to delineate, reconstruct and visualize patient organs and pathologies in 3D before surgery from medical imaging (Myrian© from Intrasense, Ziostation© from Ziosoft, Synapse© Vincent from Fujinon, Iqqa® Liver from Edda Technology, ScoutTM Liver from Pathfinder). The obtained virtual patient can then be used to facilitate or optimize diagnosis or surgical planning. When coupling preoperative information with intraoperative information, it is also possible to develop guidance software based on Augmented Reality (AR). AR displays the 3D model of the patient and a 3D modeling of instruments with overlay of the real surgical video, thus augmenting the real view with virtual information. The patient becomes then virtually transparent in the surgeon’s view so that he/she can locate vessels and tumors that are not directly visible and that he/she could previously only perceive through touching. There is currently no commercial solution offering intraoperative assistance guiding surgeons through the use of such a modeling. Research work in this field either focuses on hepatic tumor puncture (7-13), or is based on the intraoperative use of 2D ultrasonography on the liver (14-18). In the first case they generally use a rigid registration method, thus supposing that the patient remains in an identical captured position as during the acquisition before surgery. In the second case, they can only display the position of structures that are visible on the 2D ultrasonography image.
In order to overcome these limits, we have developed a comprehensive solution with the objective of simplifying the entire procedure. The first step proposes a new automated delineation method of organs and pathological structures. We have then associated a surgical planning software developed and optimized for surgical use and not for radiologic use. Finally, we have developed an interactivity-based intraoperative augmented reality assistance software. This method provides efficient and accurate augmented reality from an endoscopic or external camera, but within a limited field of view since precise global registration in a broader field of view is impossible with the proposed method. After the presentation of the first results demonstrating the feasibility of this approach, we will conclude by proposing future perspectives in order to overcome the remaining limits.
Methods
Most research studies on the liver use CT medical images taken after injection of a contrast medium at portal time (60 seconds after injection). Image dimensions however greatly vary, mainly in the craniocaudal axis (Z axis of the image). Interactive methods generally have an important slice thickness, i.e., 5 mm. Automated methods have a smaller thickness, 2 mm, and therefore have a larger number of slices. Our work relies on CT images taken at portal time with a 2 mm distance between slices. The difference between that distance and pixel size (about 0.6 mm), called anisotropy, complicates any 3D processing. This anisotropy, which can be noted in almost all existing research, sometimes makes 3D data processing more complex and requires taking that difference into account.
From these images, the three steps of our method can be applied on a standard computer, including laptops, with sufficient memory and a 3D graphic card. Experimentations have thus been conducted and validated efficiently on various different computers:
- 3D patient modeling has been done with VR-Anat©2011 on an Apple iMac with IntelCore i7 860 and 8 Go RAM, ATI Radeon 5750 with 1G memory and running under Mac OSX 10.6 Snow-Leopard; and with a Sony Viao computer with Intel® Core™ i7 CPU à 2.67 GHz fitted with 8 Go RAM, Nvidia GeForce GT 330 M and running under Windows 7 64 bits.
- Preoperative patient visualization and virtual surgical planning have been done on various computers running under MacOS 9 and 10, Windows XP, Vista and 7, and Linux debian and Ubuntu. They were standard desktop computers and laptops fitted with at least 2 Go RAM and an OpenGL compatible graphic card (Intel® Graphics Media Accelerator HD integrated, Nvidia GeForce Graphics processors and ATI Radeon Graphics processors).
- Intraoperative augmented reality assistance has been done with a Sony Viao laptop with Intel® Core™ i7 CPU 2.67 GHz and 8 Go RAM, Nvidia GeForce GT 330 M and running under Windows 7 64 bits.
So as not to develop several incompatible solutions, we have decided to develop a common architecture on which all our software tools are based: FrameWork for Software Process Line (FW4SPL). FW4SPL is a set of libraries programmed in C++ (object oriented programming language), open (source code available online) and multiplatform (Windows, Linux and MacOs). FW4SPL has been specifically designed for the development of computer-assisted surgical software. The resulting solution features many advantages (19): eased integration, portability, scalability, fast and robust development of new software, etc.
3D patient modeling
There are many methods for medical image processing proposing liver segmentation. Several authors have proposed to delineate the liver contour with automated methods. Some use deformable 3D models either to directly delineate structures (20) or to improve a result from a previous delineation method (21). Other techniques use thresholding and so-called “mathematical morphology” operators in order to carry out that delineation (22,23) or sometimes more complex operators (24). In patients presenting multiple small tumors, methods relying on deformable models seem more efficient. In contrast, when patients have an atypically-shaped liver that varies too much from the standard hepatic shape, the other methods usually provide better results. Finally, and this for any method, patients with large hypodense tumors on the liver border will fault the various methods. In this case, a manual method is preferred because the operator will be able to get around the problem. Furthermore, all these methods remain limited to liver delineation without taking delineation of neighboring organs into account or without delineating them, even if knowing organ location and shape could be useful and sometimes necessary during surgery. To overcome these limits, several research studies proposed to include hepatic segmentation in a more global segmentation of neighboring organs (25-30). Generally, they rely on a probabilistic atlas (3D cartography of image areas where each organ should be) computed from a database of real clinical cases. These methods are thus not able to tackle unusual shapes that are not represented in the image database of the atlas.
To overcome these limits, we propose a new method based on hierarchical segmentation of visible organs, from the easiest to the most complex one (31,32). The first step detects and delineates respectively skin, lungs, aorta, kidneys, spleen and bones. To do so, we translate into constraints the localization and shape knowledge and rules that are used by radiologists and surgeons when they analyze an image. These constraints are then applied to basic mathematical functions in image processing. This way, we translate organ density into thresholding, visualized contrast into gradient, number and shape into topology operators and mathematical morphology operators. From that segmentation, delineated organs are then deleted from the initial image, so as to reduce the image, becoming a smaller image containing organs that have not yet been segmented. Moreover, segmented organs are used to extract useful information (in particular localization information) for the segmentation of not yet segmented organs. The second step then delineates veins, liver and its internal tumors through automatic image density analysis coupled with shape analysis (32-34).
Finally, the last step delineates the anatomical segmentation of the liver from the portal vein. For many years, the anatomical segmentation of the liver has been debated. Couinaud’s segmentation is currently the reference used by most surgeons but it is criticized by several authors who highlight its limits, or even errors of that segmentation. Couinaud himself (35) has described those topographic anomalies. He demonstrated on 111 cases that there are incoherencies between vascular topology and segment topography which could be corrected through the use of our 3D modeling and segmentation software developed in 2001 (32). It guarantees topologic efficiency of the final segmentation and adds no artificial topographic restriction in order to avoid the limits or errors of Couinaud’s segmentation.
All these algorithms and image processing have been integrated into a new software, VR-Anat©IRCAD 2011 that also adds an interactive process making potential manual corrections possible in case of failure or inaccuracy of the automatic process. In parallel, like other existing services (MeVis Distant Services AG, PolyDimensions GmbH, Edda technology, 3DR Laboratories), we have opened an online service called “Visible Patient” that provides the 3D modeling of patients from their medical imaging sent via Internet in DICOM format. In its validation phase, this service is free of charge and is limited to partner hospitals (Nouvel Hôpital Civil in Strasbourg, France; Cavell Hospital in Brussels, Belgium; University Hospital in Montreal, Canada and Show Chwan Memorial Hospital in Changhua, Taiwan).
Virtual surgical planning software
No 3D modeling can be used without visualization software. As we have previously mentioned it, usual software tools have mainly been developed for radiological applications and do not run under all exploitation systems. To solve this problem, we have developed several software tools from our FW4SPL architecture.
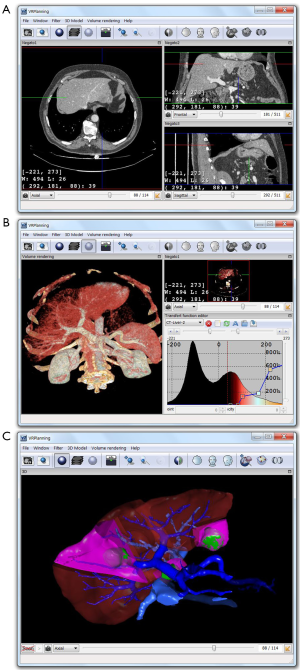
A first image visualization software (VR-Render ©IRCAD2010) working under Windows, Linux and MasOS has thus been developed for radiologists. It allows to visualize images in various formats, such as DICOM, InrImage, Jpeg, Vtk and FwXML in 2D (frontal, sagittal and axial view) or 3D slices thanks to direct volume rendering. Like all volume rendering systems available on workstations in medical imaging departments, it requires a transfer function to set the parameters of the 3D view. Several automatic rendering functions have been integrated for CT images so as to ease the use of the software. This allows to visualize the medical image in an axial, sagittal and frontal plane (Multi Planar Rendering view, or MRP) in 3D in overlay of the 3D patient modeling. The VR-Render software, freely available (www.ircad.fr/softwares/vr-render/Software.php), has been downloaded more than 20,000 times and can be downloaded on more than 15 websites.
From the same FW4SPL architecture, we have then developed a new optimized version of VR-Render dedicated to surgeons, easier to use thanks to an optimized user-friendly interface. VR-Render WeBSurg Limited Edition©IRCAD2010 is freely available on WeBSurg (http://www.websurg.com/softwares/vr-render/) since January 2010 and has been downloaded and used by more than 8,000 users. This educational website offers furthermore a range of anonymous clinical cases including their operative video. This allows for an efficient learning of its use illustrated by specific clinical cases.
From VR-Render WLE, we have finally developed a surgical planning software for virtual resection of the liver or any other organ. Compared with our previous 3D virtual surgical planning software (3D VSP) developed in 2002 (36), VR-Planning©IRCAD2010 includes all VR-Render WLE functions (2D DICOM viewer and direct volume rendering). It also gives the opportunity of cutting several liver areas (i.e., several topological components) whereas 3D VSP only permitted to cut two components (Figure 2). Multi-segmentectomy is thus possible. A second important improvement is the automatic computation of the percentage of remaining liver after resection (FLR, Future Liver Remain) whereas 3D VSP only provided the volume.
Intraoperative assistance
Preoperative surgical planning can mainly improve the efficiency of minimally invasive surgery procedures thanks to better learning of patient anatomy and real preoperative training. This use is however not sufficient to guarantee that this virtual surgical procedure will be reproduced for real. Such an improvement can be provided by intraoperative use of virtual reality via the concept of AR. Indeed, AR consists in superimposing the preoperative 3D patient modeling onto the real intraoperative view of the patient. It offers a view in transparency of patients and can also guide surgeons thanks to the virtual visualization of their real surgical tools, which are tracked throughout the procedure.
Existing methods do currently not allow for an accurate and efficient superimposition of the preoperative 3D modeling in real-time, mainly due to organ displacements and deformations during the surgical procedure compared with position and shape of these organs in the preoperative image. To overcome these limits, we propose a novel approach consisting in an interactive process called “Interactive Augmented Reality” (IAR). This method is an extension of our method described in 2004 (37) and developed for adrenal tumors. Our first method was based on IAR done from an external view of the patient. This new method does not require external registration, IAR being directly done from the camera view (laparoscopic view for minimally invasive procedures and external view for open surgery). The method consists in visualizing simultaneously, on a same screen, the intraoperative video view of the patient and the preoperative 3D modeling, and in manually changing position, scale and orientation of the 3D model so that visible landmarks coincide both on the 3D model and the real view of the patient. So that it works in real-time, manipulation of the 3D model, allowing to register it onto the real view, has been simplified as much as possible with a mere mouse control. For hepatic surgery, users will rely on visible anatomical landmarks, such as liver edges, umbilical fissure, suspensory ligaments and gallbladder.
This method is thus strongly visual, implying good hand-eye coordination. The main advantage is the opportunity to correct the registration at any time through real-time interaction on the 3D model. The user has to track movements of visible landmarks in the image and adjust in the same way the position of the 3D model in order to follow these movements. Furthermore, thanks to virtual resection tools of VR-Planning, the user can modify organ topology during the surgical intervention by virtually resecting that organ.
Results
We have validated our various processes in several preclinical and clinical validations. First of all, in the framework of the European Project PASSPORT, we have set up several free cooperation projects with distant university hospitals (Brussels, Montreal and Strasbourg) in order to offer the 3D modeling service named “Visible Patient” (www.visiblepatient.eu). The online service has done 769 clinical cases between January 2009 and June 2013 including cirrhotic liver, focal nodular hypertrophy, metastasis, HCC, cholangio-carcinoma and cyst (see Figure 3). 3D modeling was realized by radiology technicians with VR-Anat software without any external assistance and without any information on the patient (anonymous data with no indication on pathology). Images were always delivered in time allowing the surgical team to use VR-planning preoperatively.
Among this data, a first study was conducted by Dr. André Bégin at the CHUS of Sherbrooke and Dr. Réal Lapointe and Dr. Franck Vanderbrouck at the CHUM St-Luc of Montreal in order to determine the modeling quality provided by the Visible Patient service. From 2009 to 2011, 43 patients have been enrolled in the two surgical departments, the majority having liver cancer (37/43), mostly from colorectal origin (30/37), with an average age of 56 (range, 19-74). A comparison between manual delineation and the Visible Patient service result shows a really good correlation in the total liver volume (0.989), for the tumor volume (0.989) as well as the FLR (0.917). This study clearly demonstrated that the Visible Patient service is accurate and reliable to determine the total and FLR volumes.
A second validation was conducted by Prof. Jean-Jacques Houben at the digestive surgical department of the Cavell Hospital in Brussels in order to determine the potential clinical benefit of the Visible Patient service including the VR-Planning software. From 2009 to 2011, 48 patients have been admitted in this department for major liver pathologies. Seventeen patients among these 48 patients were sent to the Visible Patient service for 3D modeling, due to a clearly hypothetical R0 curative resection, patients not being directly eligible for surgery with the usual analysis. Eleven patients had liver metastases, three HCC and three major cysts. The 3D models were always provided by Visible Patient before any therapeutic decision, providing thus precise 3D vision of patient-specific anatomy and pathology. Among the 17 selected patients, 13 procedures were modified and ten benefited from a safe hepatectomy and R0 resection confirmed by PET scan, CEA drop and clinical outcome. Thus, thanks to 3D modeling and the associated VR-Planning software, 27% of surgical procedures for liver pathology have been modified in comparison with conventional surgical planning without such tools.
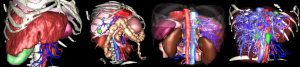
A third evaluation was conducted in our surgical department to test the efficiency of IAR. This technique has been applied on 40 surgical oncological procedures for liver, adrenal gland, pancreas, and parathyroid tumor resection. The last ones were realized for liver surgery by superimposing the virtual view onto the master control system view of the Da Vinci robot (Figure 4). Superimposition has been possible for each operation. Vascular control also showed the efficiency of the system despite the interaction required to make it work. An optimized positioning of tools has also been realized thanks to the use of AR.

Discussion
We have developed a new computer-assisted surgical procedure based on patient-specific geometrical and anatomical modeling. Combined with surgical planning software dedicated to liver surgery it offers new possibilities allowing to improve hepatic surgery. Used intraoperatively it can guide surgeons by providing them an augmented reality view. If the 3D modeling process under an online service seems clearly efficient, intraoperative assistance will need more validation to prove a clinical benefit that seems evident. However, based on a user-dependent interaction, the proposed method will remain limited and will have to be replaced in the future by integrating real-time deformable models of organs. Future solutions will thus certainly combine predictive simulation and real-time medical image analysis in order to overcome these limitations. To be efficient, patient-specific modeling will have to integrate more information than the geometric model only. Mechanical properties, functional anatomy and biological modeling will gradually improve the quality of simulation and prediction that, combined with intra-operative image analysis, will provide the necessary accuracy.
This research represents the first essential phase for surgical gesture automation, which will allow for the reduction of surgical mistakes. Indeed, surgical intervention planning will allow in a first step to identify the unnecessary or imperfect surgical movements. Then, these movements will be transmitted to a surgical robot which, using augmented reality and visual servoing, will be able to precisely reproduce the surgeon’s optimized gestures. Tomorrow’s surgery is on its way.
Acknowledgements
We would like to thank all our clinical partners, and more specifically Professor Jean-Jacques Houben at the Cavell Hospital in Brussels, Dr. André Bégin at the CHUS of Sherbrooke, Dr. Réal Lapointe and Dr. Franck Vanderbrouck at the CHUM St-Luc of Montreal, and Professor Catherine Roy at the New Civil Hospital in Strasbourg.
Disclosure: This work is a part of the eHealth project PASSPORT of the European Commission in the 7th Framework Programme, funded by the ICT program.
References
- Couinaud C. Liver anatomy: portal (and suprahepatic) or biliary segmentation. Dig Surg 1999;16:459-67. [PubMed]
- Bismuth H. Surgical anatomy and anatomical surgery of the liver. World J Surg 1982;6:3-9. [PubMed]
- Lamadé W, Glombitza G, Fischer L, et al. The impact of 3-dimensional reconstructions on operation planning in liver surgery. Arch Surg 2000;135:1256-61. [PubMed]
- Radtke A, Sgourakis G, Sotiropoulos GC, et al. Territorial belonging of the middle hepatic vein in living liver donor candidates evaluated by three-dimensional computed tomographic reconstruction and virtual liver resection. Br J Surg 2009;96:206-13. [PubMed]
- OsiriX Imaging Software, Available online: www.osirix-viewer.com
- Render VR, Websurg LE, Available online: www.websurg.com/softwares/vr-render. VR-Render, Available online: www.ircad.fr/softwares/vr-render
- Wacker FK, Vogt S, Khamene A, et al. An augmented reality system for MR image-guided needle biopsy: initial results in a swine model. Radiology 2006;238:497-504. [PubMed]
- Fischer GS, Deguet A, Csoma C, et al. MRI image overlay: application to arthrography needle insertion. Comput Aided Surg 2007;12:2-14. [PubMed]
- Fichtinger G, Deguet A, Masamune K, et al. Image overlay guidance for needle insertion in CT scanner. IEEE Trans Biomed Eng 2005;52:1415-24. [PubMed]
- Lasowski R, Benhimane S, Vogel J, et al. Adaptive visualization for needle guidance in RF liver ablation: taking organ deformation into account. roc. SPIE 6918, Medical Imaging 2008 Visualization, Image-guided Procedures, and Modeling 2008;69180A: [PubMed]
- Nicolau SA, Pennec X, Soler L, et al. An augmented reality system for liver thermal ablation: design and evaluation on clinical cases. Med Image Anal 2009;13:494-506. [PubMed]
- Khan MF, Dogan S, Maataoui A, et al. Navigation-based needle puncture of a cadaver using a hybrid tracking navigational system. Invest Radiol 2006;41:713-20. [PubMed]
- Yaniv Z, Cheng P, Wilson E, et al. Needle-based interventions with the image-guided surgery toolkit (IGSTK): from phantoms to clinical trials. IEEE Trans Biomed Eng 2010;57:922-33. [PubMed]
- Feuerstein M, Reichl T, Vogel J, et al. Magneto-optical tracking of flexible laparoscopic ultrasound: model-based online detection and correction of magnetic tracking errors. IEEE Trans Med Imaging 2009;28:951-67. [PubMed]
- Nakamoto M, Hirayama H, Sato Y, et al. Recovery of respiratory motion and deformation of the liver using laparoscopic freehand 3D ultrasound system. Med Image Anal 2007;11:429-42. [PubMed]
- Shekhar R, Dandekar O, Bhat V, et al. Live augmented reality: a new visualization method for laparoscopic surgery using continuous volumetric computed tomography. Surg Endosc 2010;24:1976-85. [PubMed]
- Hostettler A, George D, Rémond Y, et al. Bulk modulus and volume variation measurement of the liver and the kidneys in vivo using abdominal kinetics during free breathing. Comput Methods Programs Biomed 2010;100:149-57. [PubMed]
- Hostettler A, Nicolau SA, Rémond Y, et al. A real-time predictive simulation of abdominal viscera positions during quiet free breathing. Prog Biophys Mol Biol 2010;103:169-84. [PubMed]
- Fasquel J, Chabre G, Zanne P, et al. A role-based component architecture for computer assisted interventions: illustration for electromagnetic tracking and robotized motion rejection in flexible endoscopy. MIDAS Journal, Systems and Architectures for Computer Assisted Interventions 2009, Available online: http://www.midasjournal.org/browse/publication/648.
- Lamecker H, Lange T, Seebass M, et al. Automatic segmentation of the liver for preoperative planning of resections. Stud Health Technol Inform 2003;94:171-3. [PubMed]
- Seong W, Kim EJ, Park JW. Automatic segmentation technique without user modification for 3D visualization in medical images. Lecture Notes in Computer Science Volume 2005;3314:595-600.
- Soler L, Delingette H, Malandain G, et al. Fully automatic anatomical, pathological, and functional segmentation from CT scans for hepatic surgery. Comput Aided Surg 2001;6:131-42. [PubMed]
- Lin SJ, Jeong YY, Ho YS. Automatic liver segmentation for volume measurement in CT Images. Journal of Visual Communication and Image Representation 2006;17:860-75.
- Chemouny S, Henry H, Masson B, et al. Advanced 3D image processing techniques for liver and hepatic tumor location and volumetry. Proc SPIE 3661, Medical Imaging 1999: Image Processing, 761; doi:. [PubMed]
- Furst JD, Susomboom R, Raicu DS. Single organ segmentation filters for multiple organ segmentation. Conf Proc IEEE Eng Med Biol Soc 2006;1:3033-6. [PubMed]
- Whitney BW, Backman NJ, Furst JD, et al. Single click volumetric segmentation of abdominal organs in Computed Tomography images. Medical Imaging: Image Processing. Proceedings of the SPIE 2006;6144:1426-36.
- Zhou Y, Bai J. Atlas based automatic identification of abdominal organs. Proc SPIE 5747, Medical Imaging 2005: Image Processing, 1804; doi:. [PubMed]
- Kitasaka T, Ogawa H, Yokoyama K, et al. Automated extraction of abdominal organs from uncontrasted 3D abdominal X-Ray CT images based on anatomical knowledge. Journal of Computer Aided Diagnosis of Medical Images 2005;9:1-14.
- Camara O, Colliot O, Bloch I. Computational modeling of thoracic and abdominal anatomy using spatial relationships for image segmentation. Real-Time Imaging 2004;10:263-73.
- Park H, Bland PH, Meyer CR. Construction of an abdominal probabilistic atlas and its application in segmentation. IEEE Trans Med Imaging 2003;22:483-92. [PubMed]
- Soler L, Nicolau S, Hostettler A, et al. eds. Computer Assisted Digestive Surgery, Computational Surgery and Dual Training. US Ed: Springer, 2010;139-53.
- Mutter D, Soler L, Marescaux J. Recent advances in liver imaging. Expert Rev Gastroenterol Hepatol 2010;4:613-21. [PubMed]
- Ruskó L, Bekes G, Fidrich M. Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med Image Anal 2009;13:871-82. [PubMed]
- Heimann T, Meinzer HP, Wolf I. A statistical deformable model for the segmentation of liver CT volumes. In Proc MICCAI Workshop on 3D Segmentation in the Clinic: a Grand Challenge 2007:161-6.
- Couinaud C. Errors in the topographic diagnosis of liver diseases. Ann Chir 2002;127:418-30. [PubMed]
- Koehl C, Soler L, Marescaux J. A PACS Based Interface for 3D Anatomical Structures Visualization and Surgical Planning. SPIE proceeding 2002;4681:17-24.
- Marescaux J, Rubino F, Arenas M, et al. Augmented-reality-assisted laparoscopic adrenalectomy. JAMA 2004;292:2214-5. [PubMed]


