Published online Nov 21, 2019. doi: 10.3748/wjg.v25.i43.6451
Peer-review started: August 19, 2019
First decision: September 10, 2019
Revised: September 17, 2019
Accepted: October 17, 2019
Article in press: October 17, 2019
Published online: November 21, 2019
Because of the powerful abilities of self-learning and handling complex biological information, artificial neural network (ANN) models have been widely applied to disease diagnosis, imaging analysis, and prognosis prediction. However, there has been no trained preoperative ANN (preope-ANN) model to preoperatively predict the prognosis of patients with gastric cancer (GC).
To establish a neural network model that can predict long-term survival of GC patients before surgery to evaluate the tumor condition before the operation.
The clinicopathological data of 1608 GC patients treated from January 2011 to April 2015 at the Department of Gastric Surgery, Fujian Medical University Union Hospital were analyzed retrospectively. The patients were randomly divided into a training set (70%) for establishing a preope-ANN model and a testing set (30%). The prognostic evaluation ability of the preope-ANN model was compared with that of the American Joint Commission on Cancer (8th edition) clinical TNM (cTNM) and pathological TNM (pTNM) staging through the receiver operating characteristic curve, Akaike information criterion index, Harrell's C index, and likelihood ratio chi-square.
We used the variables that were statistically significant factors for the 3-year overall survival as input-layer variables to develop a preope-ANN in the training set. The survival curves within each score of the preope-ANN had good discrimination (P < 0.05). Comparing the preope-ANN model, cTNM, and pTNM in both the training and testing sets, the preope-ANN model was superior to cTNM in predictive discrimination (C index), predictive homogeneity (likelihood ratio chi-square), and prediction accuracy (area under the curve). The prediction efficiency of the preope-ANN model is similar to that of pTNM.
The preope-ANN model can accurately predict the long-term survival of GC patients, and its predictive efficiency is not inferior to that of pTNM stage.
Core tip: We established an artificial neural network model before surgery that can predict the long-term survival of patients with gastric cancer, and its predictive efficiency is not inferior to that of pathological TNM stage.
- Citation: Que SJ, Chen QY, Qing-Zhong, Liu ZY, Wang JB, Lin JX, Lu J, Cao LL, Lin M, Tu RH, Huang ZN, Lin JL, Zheng HL, Li P, Zheng CH, Huang CM, Xie JW. Application of preoperative artificial neural network based on blood biomarkers and clinicopathological parameters for predicting long-term survival of patients with gastric cancer. World J Gastroenterol 2019; 25(43): 6451-6464
- URL: https://www.wjgnet.com/1007-9327/full/v25/i43/6451.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i43.6451
Gastric cancer (GC) is one of the five most common malignant tumors in the world, and it is the third leading cause of death in cancer patients[1]. Despite the progress in the diagnosis and treatment of GC, the prognosis of GC patients is still very poor, especially the long-term survival of advanced GC patients, for which the 5-year survival rate is only 10%[2]. Most scholars have focused on how to accurately distinguish the stages of GC in patients. The improved postoperative American Joint Commission on Cancer (AJCC) TNM staging system based on pathological examination is currently the most important and recognized prognostic staging system for GC[3-5]. However, this scoring system needs to be performed based on the pathological analysis of tumor specimens and examination of lymph nodes after surgery, which cannot provide a reference for preoperative treatment and consultation. Recently, the treatment for GC has gradually changed from simple surgical treatment to comprehensive treatment with the core being surgical treatment. Accurate preoperative tumor assessment providing a reasonable individualized treatment for GC patients is the key to improving the prognosis of patients with GC. However, the single traditional index, namely, the clinical TNM (cTNM) staging system based on the imaging examination, does not show the ideal accuracy of preoperative tumor assessment. Therefore, it has been a challenge in clinical work to explore the markers and methods for preoperative accurate tumor assessment.
In recent years, increasingly more scholars have believed that the inflammation indexes are relevant to the survival of cancer patients[6,7]. Some scholars have reported that inflammatory cells may promote tumor growth and progression of the disease, rather than produce an effective host antitumor response. Consequently, the inflammatory biomarkers in peripheral blood are potential predictors of cancer prognosis[8-11]. It has been reported that several indexes from peripheral blood, such as the neutrophil-lymphocyte ratio (NLR), platelet-lymphocyte ratio (PLR), and albumin-globulin ratio (AGR), have a powerful impact on the prognosis of GC[8,11-14]. The prognostic nutrition index (PNI) based on lymphocyte and albumin levels is a feasible parameter to reflect the immune and nutritional status of patients with a malignant tumor and is regarded as a good index to evaluate the prognosis of patients with GC[3,15]. Although the previously established inflammatory models could predict the prognosis of patients with GC, their accuracy was still not satisfactory. Furthermore, many pathophysiological processes of malignant tumors are nonlinear processes, which cannot be well reflected through traditional linear analysis methods. Many studies have shown that artificial neural networks (ANNs) can deal with the nonlinear statistical relationship better than the traditional analysis methods, including studies on the prognosis of various cancers[16-18]. It has been reported that ANNs had a more accurate prognostic ability than TNM staging in patients with breast and rectal cancers[19]. However, there is no study on the relationship between a preoperative ANN (preope-ANN) and the prognosis in GC patients. Therefore, this study combined the preoperative blood biomarkers and preoperative tumor data to establish an ANN model in order to build a reliable preoperative prediction system that can achieve the same effect of postoperative TNM staging. The aim of this study was to evaluate the prognosis of patients with GC and to provide a reasonable individualized treatment plan for patients.
The clinicopathological data of 1860 GC patients treated at the Department of Gastric Surgery, Fujian Medical University Union Hospital from January 2011 to April 2015 were analyzed retrospectively. The inclusion criteria were that all patients were diagnosed with GC and accepted radical surgery at our center. The exclusion criteria were as follows: (1) The general clinical data were incomplete, including age, sex, body mass index (BMI), American society of anesthesiologists (ASA) score, carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA199), alpha fetoprotein (AFP), NLR, PLR, AGR, PNI, preoperative complications, tumor size, primary site, clinical T stage (cT), and clinical N stage (cN); (2) The follow-up data were missing; and (3) Patients receiving neoadjuvant chemotherapy. A total of 1608 patients were included (Supplementary Figure 1). The study was approved by the Ethics Committee of Fujian Medical University Union Hospital.
All patients underwent routine preoperative examinations, including upper gastrointestinal endoscopy and upper gastrointestinal angiography, to assess tumor location. Preoperative clinical staging was assessed using chest radiography, total abdominal computed tomography (CT), abdominal ultrasound, and, if necessary, PET-CT, and bone scans. The 8th edition of the AJCC tumor staging system was used for cTNM and pathological TNM (pTNM) staging (Supplementary Figure 1). The overall survival (OS) time was the time from the operation to the last follow-up time or death.
All patients were randomly divided into a training set (n = 1104, 70%) and a testing set (n = 504, 30%), and the testing set was separated for evaluation of the final model. The training data were used five times for cross-validation to optimize the neural network. Finally, the separated data of the testing set were used to evaluate the model.
Routine blood biochemistry and tumor markers were assessed within 1 wk before the operation. Using X-tile software, the optimal critical values of the NLR for OS were 1.91 and 3.87 (P < 0.05). According to optimal critical values, the patients were divided into three groups as follows: Low NLR group (NLR ≤ 1.91, n = 632), middle NLR group (1.91 < NLR ≤ 3.87, n = 753), and high NLR group (NLR > 3.87, n = 223). Similarly, the patients were divided into a low PLR group (PLR ≤ 89.5, n = 258), middle PLR group (89.5 < PLR ≤ 162.3, n = 777), and high PLR group (PLR > 162.3, n = 573). According to the AGR, the patients were divided into a low AGR group (AGR ≤ 6.59, n = 474), middle AGR group (6.59 < AGR ≤ 7.81, n = 612), and high AGR group (AGR > 7.81, n = 522). Furthermore, according to the PNI, the patients were divided into a low PNI group (PNI ≤ 371.01, n = 495), middle PNI group (371.0 < PNI ≤ 430.0, n = 771), and high PNI group (PNI > 430.0, n = 342).
First, logistic univariate analysis was used to analyze 1104 cases in the training set to screen the variables that affected the 3-year survival and to determine these variable items as the input nodes of the ANN.
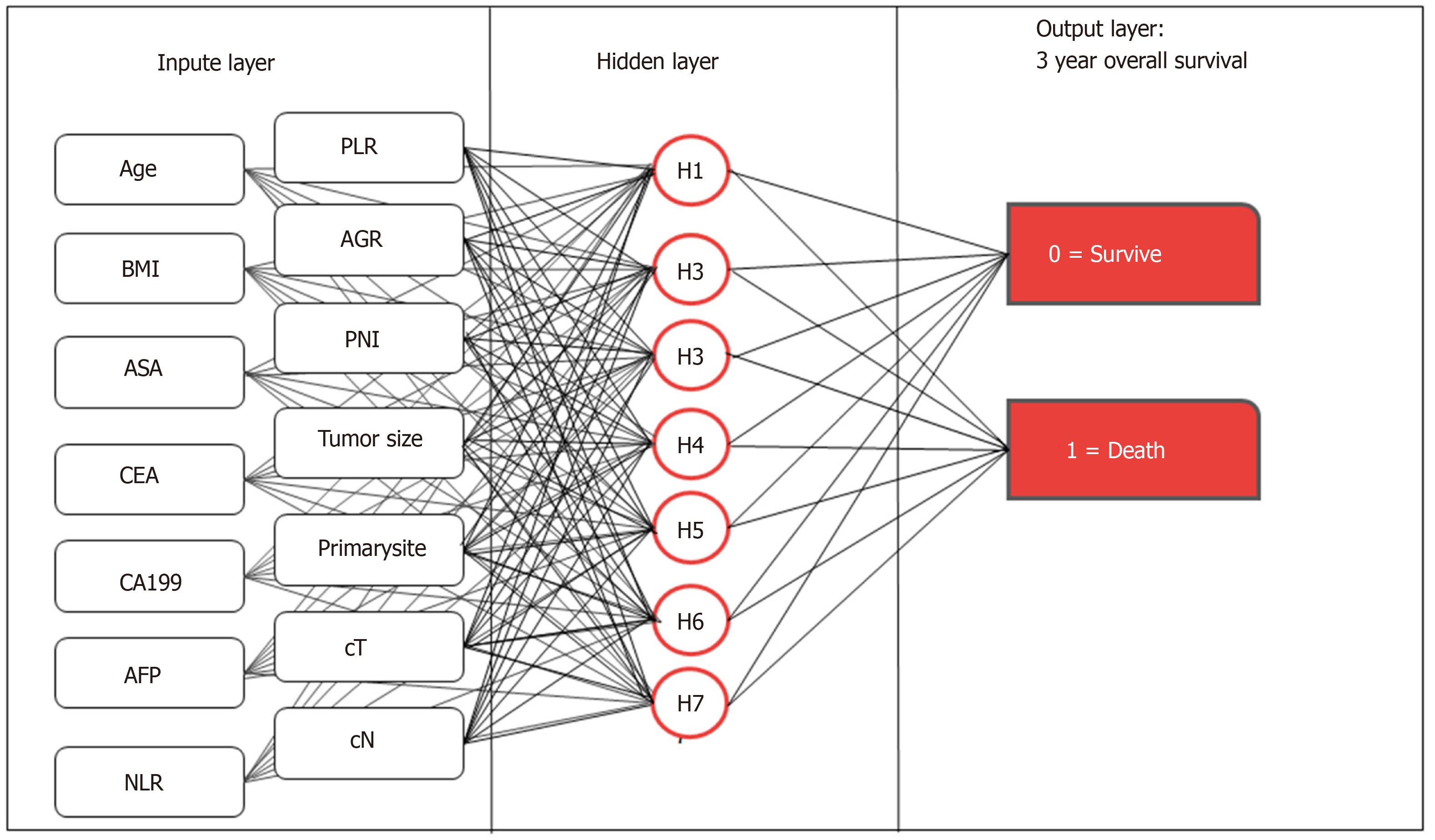
We used the multilayer perceptron to develop the preope-ANN (Figure 1). The input layer of the preope-ANN model consisted of the screened general clinicopathological data and preoperative blood biomarkers using logistic analysis and the 3-year survival condition. The ANN intelligently analyzed the 3-year survival of the patients according to the input data and generated the outcome compared with the real value. The output layer consisted of two values: (1) The patient's 3-year survival (death or survival); and (2) The continuous variables from 0 to 1, which represent the predictive probability of the outcome of the patient's 3-year survival. The network architecture of the preope-ANN model was composed of seven hidden layers, and the input layer was composed of 16 nodes (including 14 general clinical data and blood biomarkers and two survival conditions related to the 3-year OS). The hidden layer was activated by the hyperbolic tangent function, and the output layer was composed of two nodes activated by the softmax function, including the survival condition and survival probability. According to the survival probability predicted by the preope-ANN model, the patients were divided into seven subgroups: The survival probability was 0%-30% for group A, 30%-50% for group B, 50%-65% for group C, 65%-80% for group D, 80%-90% for group E, 90%-97% for group F, and 97%-100% for group G.
All data were analyzed using SPSS 20.0 (SPSS Inc. Chicago, IL, United States) and R software (version 3.5.4). The classified variables were tested using the χ2 test or Fisher's exact test. The X-tile software was used to determine the best cut-off point of the counting data. Mann-Whitney U test was used to test the receiver operating characteristic (ROC) curve and the area under the curve (AUC), and the Z test was used to compare the AUCs (MedCal software). We used Harrell’s C index to measure the discriminatory ability of the different models[4,20]. The likelihood ratio chi-square was calculated by Cox regression to measure homogeneity; a higher score means better homogeneity[21]. The Akaike information criterion (AIC) within the Cox regression model was used to compare performances between two prognostic models; smaller AIC values represent better optimistic prognostic stratification[22]. We calculated the relative likelihood of two models using the following formula: Exp {[AIC (model A) - AIC (model B)]/2}. The relative likelihood represents the probability that model A minimizes information as effectively as model B and could thus be interpreted as a P value for the comparison of both AIC values[23]. P < 0.05 was considered statistically significant.
Table 1 shows the clinical data of 1608 patients with GC. Among them, 1104 cases were in the training set, and 504 cases were in the testing set. The average age of all patients diagnosed was 60.72 years (range, 12-101 years), and the male to female ratio was 2.84:1. The proportion of patients diagnosed with stages I, II, III, and IVA disease in the cTNM system was 11.6%, 38.2%, 45.6%, and 4.6%, respectively. The proportion of patients diagnosed with stage I, II, and III disease in the pTNM system was 22.6%, 24.9%, and 52.5%, respectively.
| Variable | All patients (n = 1608) | Training set (n = 1104) | Testing set (n = 504) |
| Age (yr) | |||
| < 57 | 593 (36.9) | 413 (37.4) | 180 (35.7) |
| 57-74 | 838 (52.1) | 565 (51.2) | 273 (54.2) |
| > 74 | 177 (11.0) | 126 (11.4) | 51 (10.1) |
| Sex | |||
| Female | 412 (25.6) | 299 (27.1) | 113 (22.4) |
| Male | 1196 (74.4) | 805 (72.9) | 391 (77.6) |
| BMI | |||
| < 18.5 | 156 (9.7) | 98 (8.9) | 58 (11.5) |
| 18.5-23.5 | 960 (59.7) | 670 (60.7) | 290 (57.5) |
| > 23.5 | 492 (30.6) | 336 (30.4) | 156 (31.0) |
| CEA | |||
| < 2.8 | 932 (58.0) | 626 (56.7) | 306 (60.7) |
| 2.8-4.8 | 328 (20.4) | 226 (20.5) | 102 (20.2) |
| > 4.8 | 348 (21.6) | 252 (22.8) | 96 (19.0) |
| CA199 | |||
| < 15.0 | 1043 (64.9) | 696 (63.0) | 347 (68.8) |
| 15.0-39.2 | 348 (21.6) | 255 (23.1) | 93 (18.5) |
| > 39.2 | 217 (13.5) | 153 (13.9) | 64 (12.7) |
| AFP | |||
| < 2 | 496 (30.8) | 344 (31.2) | 152 (30.2) |
| 2-5.29 | 916 (57.0) | 631 (57.2) | 285 (56.5) |
| > 5.3 | 196 (12.2) | 129 (11.7) | 67 (13.3) |
| NLR | |||
| < 1.91 | 632 (39.3) | 422 (38.2) | 210 (41.7) |
| 1.91-3.87 | 753 (46.8) | 525 (47.6) | 228 (45.2) |
| > 3.87 | 223 (13.9) | 157 (14.2) | 66 (13.1) |
| PLR | |||
| < 89.5 | 258 (16.0) | 177 (16.0) | 81 (16.1) |
| 89.5-162.3 | 777 (48.3) | 540 (48.9) | 237 (47.0) |
| >162.3 | 573 (35.6) | 387 (35.1) | 186 (36.9) |
| AGR | |||
| < 6.59 | 474 (29.5) | 332 (30.1) | 142 (28.2) |
| 6.59-7.81 | 612 (38.1) | 426 (38.6) | 186 (36.9) |
| > 7.81 | 522 (32.5) | 346 (31.3) | 176 (34.9) |
| PNI | |||
| < 371 | 495 (30.8) | 347 (31.4) | 148 (29.4) |
| 371-430 | 771 (47.9) | 531 (48.1) | 240 (47.6) |
| > 430 | 342 (21.3) | 226 (20.5) | 116 (23.0) |
| ASA | |||
| I | 983 (61.1) | 677 (61.3) | 306 (60.7) |
| II | 561 (34.9) | 379 (34.3) | 182 (36.1) |
| III-IV | 64 (4.0) | 48 (4.3) | 16 (3.2) |
| Comorbidity | |||
| No | 1133 (70.5) | 779 (70.6) | 354 (70.2) |
| Yes | 475 (29.5) | 325 (29.4) | 150 (29.8) |
| Primary site | |||
| Lower | 671 (41.7) | 476 (43.1) | 195 (38.7) |
| Middle | 350 (21.8) | 231 (20.9) | 119 (23.6) |
| Upper | 401 (24.9) | 257 (23.3) | 144 (28.6) |
| Overlapping | 186 (11.6) | 140 (12.7) | 46 (9.1) |
| Tumor size (mm) | |||
| < 30 | 578 (35.9) | 404 (36.6) | 174 (34.5) |
| 30-60 | 711 (44.2) | 477 (43.2) | 234 (46.4) |
| > 60 | 319 (19.8) | 223 (20.2) | 96 (19.0) |
| cT | |||
| T1 | 167 (10.4) | 108 (9.8) | 59 (11.7) |
| T2 | 185 (11.5) | 135 (12.2) | 50 (9.9) |
| T3 | 429 (26.7) | 292 (26.4) | 137 (27.2) |
| T4 | 827 (51.4) | 569 (51.5) | 258 (51.2) |
| cN | |||
| N0 | 662 (41.2) | 457 (41.4) | 205 (40.7) |
| N1 | 376 (23.4) | 257 (23.3) | 119 (23.6) |
| N2 | 362 (22.5) | 245 (22.2) | 117 (23.2) |
| N3 | 208 (12.9) | 145 (13.1) | 63 (12.5) |
| Follow-up duration (mo) | 48 (3-91) | 48 (3-91) | 50 (3-89) |
In the training set, the univariate logistics regression analysis (Table 2) showed that age, sex, BMI, CEA, CA199, AFP, NLR, PLR, AGR, PNI, ASA score, tumor location, tumor size, cT stage, and cN stage were significant factors for the 3-year OS of the patients (P < 0.05 for all).
| Variable | Training set | |||
| OR | 95CI | P value | ||
| Age (yr) | ||||
| < 57 | REF | |||
| 57-74 | 0.38 | 0.25 | 0.58 | < 0.001 |
| > 74 | 0.63 | 0.42 | 0.93 | 0.020 |
| Sex | ||||
| Male | REF | |||
| Female | 0.96 | 0.72 | 1.29 | 0.782 |
| BMI | ||||
| < 18.5 | REF | |||
| 18.5-23.5 | 3.04 | 1.90 | 4.85 | < 0.001 |
| > 23.5 | 1.28 357 | 0.95 | 1.73 | 0.111 |
| CEA | ||||
| < 2.8 | REF | |||
| 2.8-4.8 | 0.33 | 0.24 | 0.45 | < 0.001 |
| > 4.8 | 0.46 | 0.31 | 0.67 | < 0.001 |
| CA199 | ||||
| < 15.0 | REF | |||
| 15.0-39.2 | 0.20 | 0.14 | 0.29 | < 0.001 |
| > 39.2 | 0.33 | 0.22 | 0.50 | < 0.001 |
| AFP | ||||
| < 2 | REF | < 0.001 | ||
| 2-5.29 | 0.78 | 0.51 | 1.18 | 0.240 |
| > 5.3 | 0.50 | 0.33 | 0.74 | 0.001 |
| NLR | ||||
| < 1.91 | REF | |||
| 1.91-3.87 | 0.32 | 0.22 | 0.48 | < 0.001 |
| > 3.87 | 0.48 | 0.33 | 0.69 | < 0.001 |
| PLR | ||||
| < 89.5 | REF | |||
| 89.5-162.3 | 0.26 | 0.16 | 0.42 | < 0.001 |
| > 162.3 | 0.59 | 0.45 | 0.78 | < 0.001 |
| AGR | ||||
| < 6.59 | REF | |||
| 6.59-7.81 | 2.11 | 1.50 | 2.98 | < 0.001 |
| > 7.81 | 1.58 | 1.13 | 2.20 | 0.007 |
| PNI | ||||
| < 371 | REF | |||
| 371-430 | 3.98 | 2.62 | 6.06 | < 0.001 |
| > 430 | 1.94 | 1.29 | 2.92 | 0.002 |
| ASA | ||||
| I | REF | |||
| II | 0.44 | 0.24 | 0.79 | 0.006 |
| III-IV | 0.53 | 0.29 | 0.97 | 0.041 |
| Comorbidity | ||||
| No | REF | |||
| Yes | 0.99 | 0.74 | 1.31 | 0.933 |
| Primary site | ||||
| Lower | REF | |||
| Middle | 0.30 | 0.20 | 0.45 | < 0.001 |
| Upper | 0.54 | 0.35 | 0.83 | 0.001 |
| Overlapping lesion of the stomach | 0.47 | 0.31 | 0.73 | < 0.001 |
| Tumor size (mm) | ||||
| < 30 | REF | |||
| 30-60 | 0.08 | 0.06 | 0.13 | < 0.001 |
| > 60 | 0.43 | 0.31 | 0.60 | < 0.001 |
| cT | ||||
| T1 | REF | |||
| T2 | 0.05 | 0.02 | 0.13 | < 0.001 |
| T3 | 0.18 | 0.11 | 0.32 | < 0.001 |
| T4 | 0.24 | 0.17 | 0.34 | < 0.001 |
| cN | ||||
| N0 | REF | |||
| N1 | 0.18 | 0.12 | 0.27 | < 0.001 |
| N2 | 0.28 | 0.18 | 0.43 | < 0.001 |
| N3 | 0.67 | 0.44 | 1.007 | 0.054 |
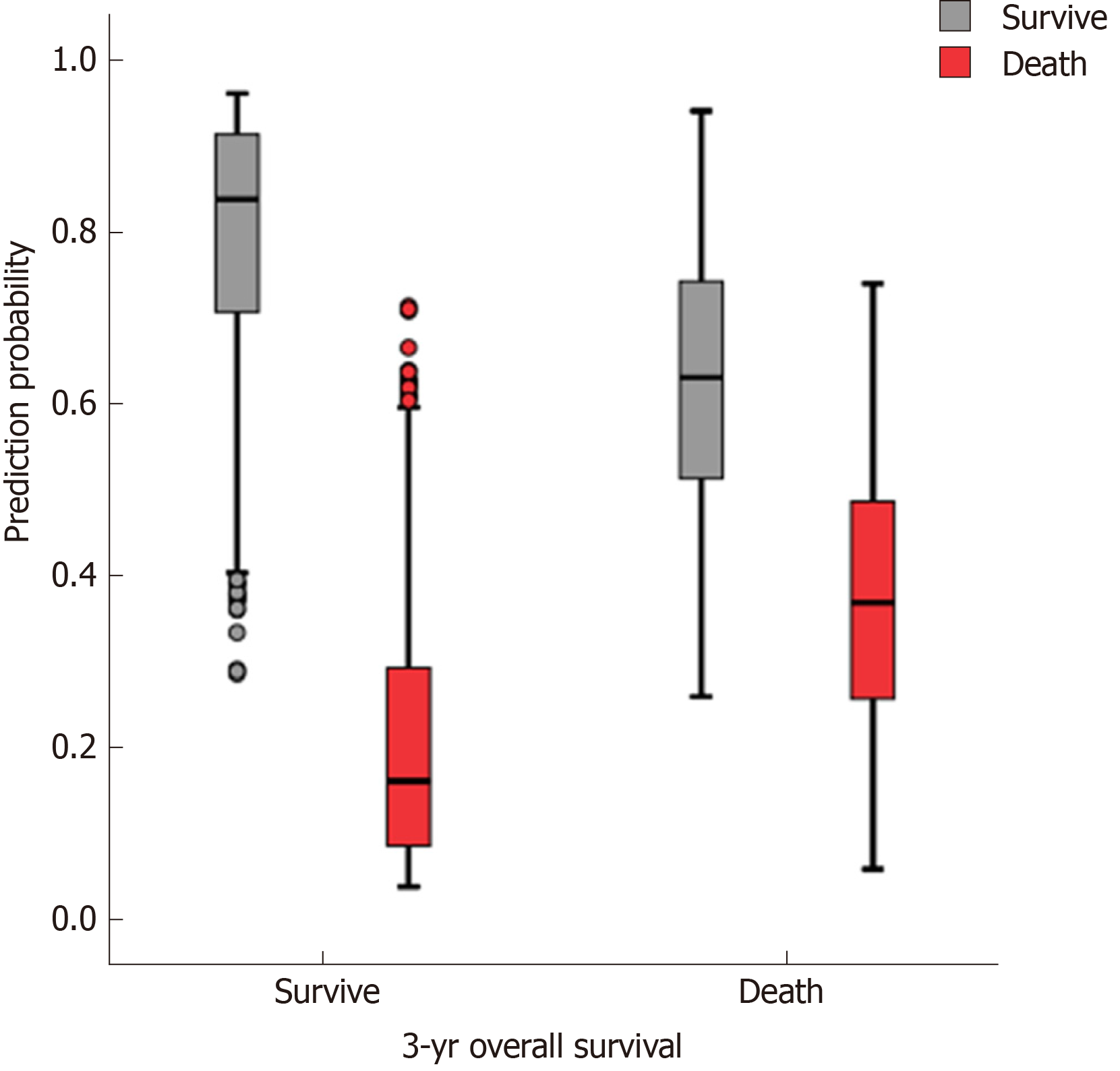
The accuracy, sensitivity, and specificity of the preope-ANN model in the training set were 77.3%, 88.5%, and 50.1%, respectively. For the preope-ANN model in the testing set, the accuracy was 75.2%, the sensitivity was 86.5%, and the specificity was 43.8% (Figure 2).
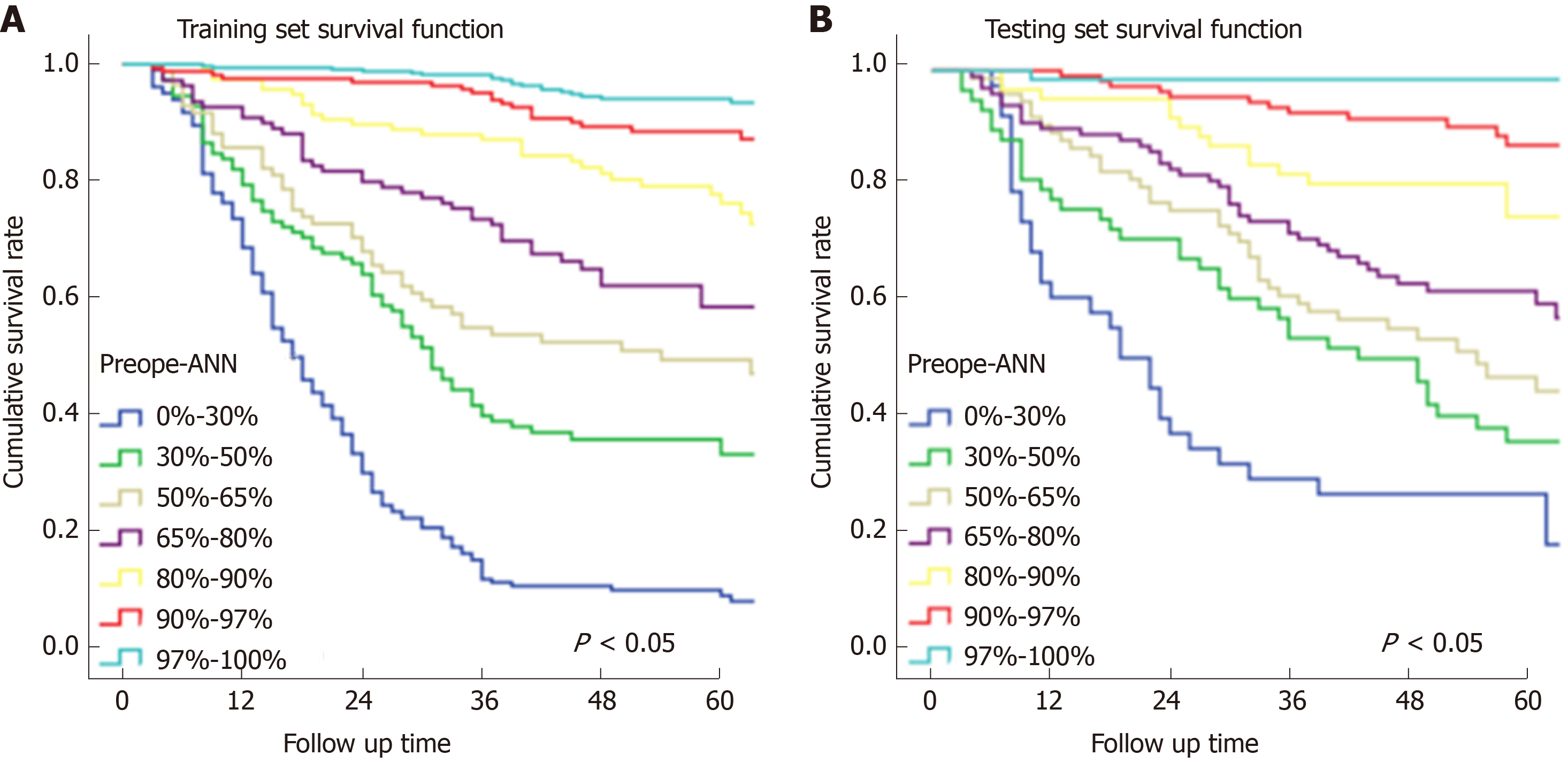
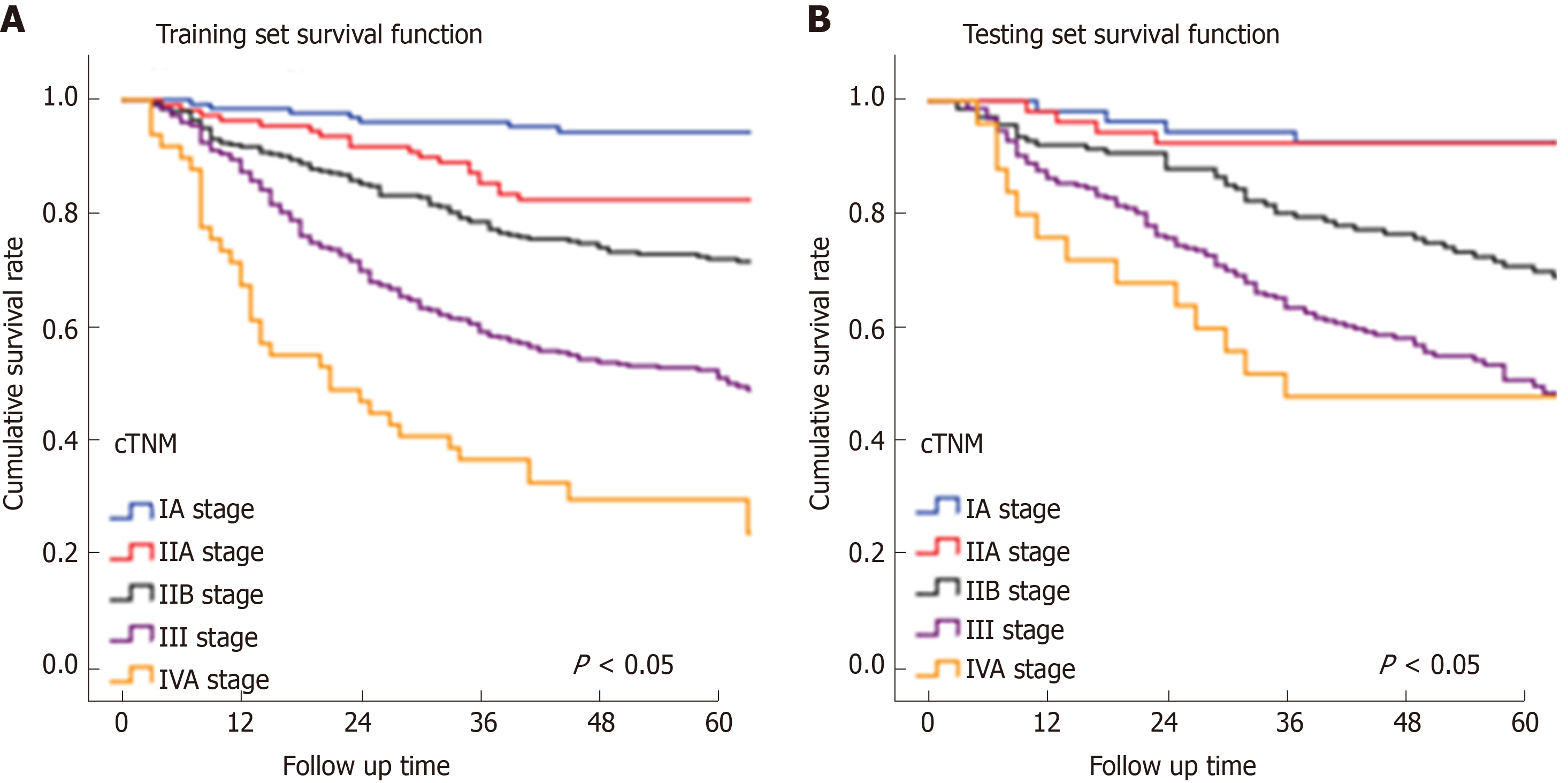
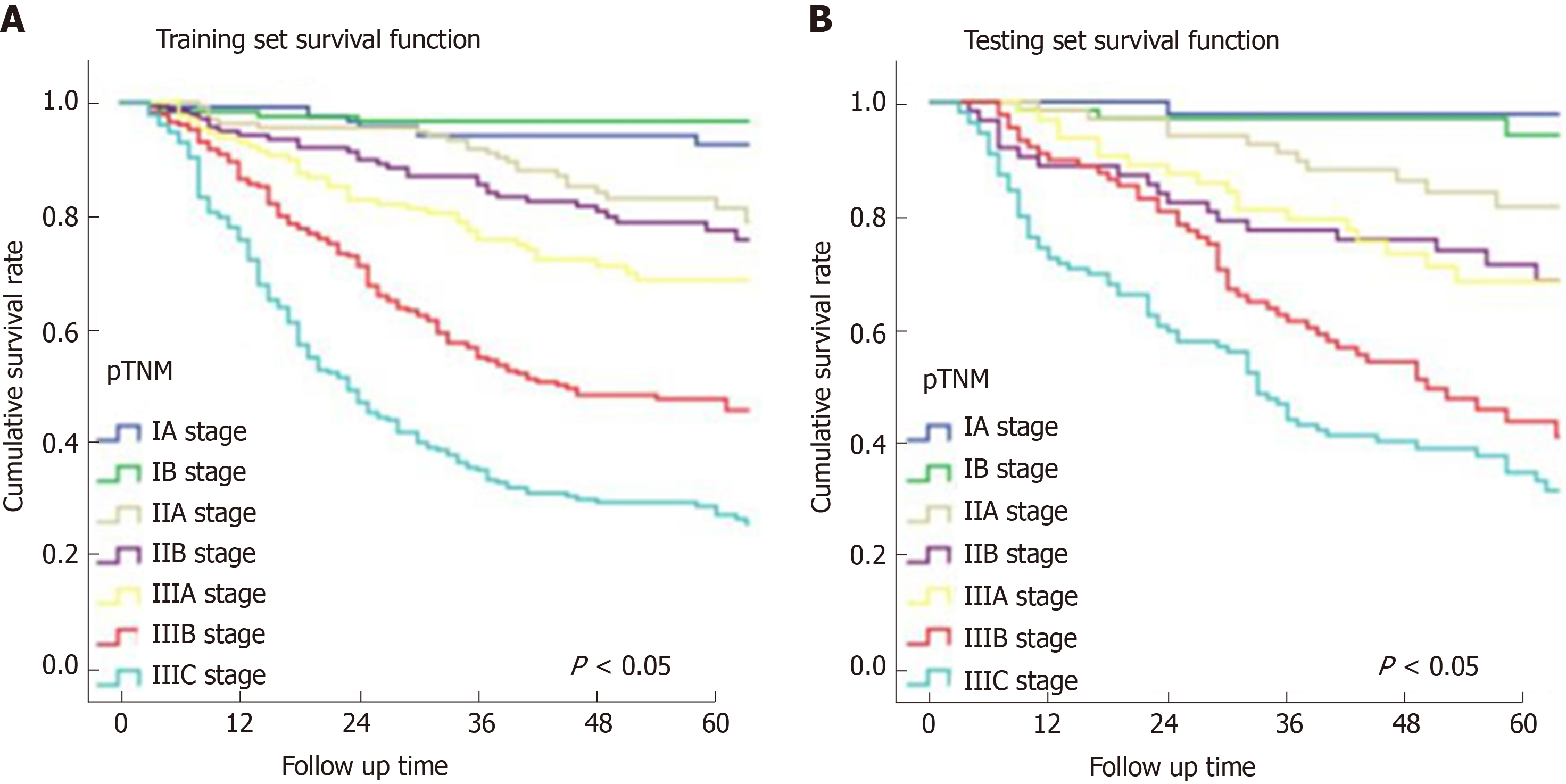
In Figure 3, the prediction results of the preope-ANN model were divided into seven subgroups. The Kaplan-Meier survival curve showed that the survival curve of each subgroup within the preope-ANN model had good discrimination in both the training and testing sets (P < 0.05). For cTNM staging, the survival curve in the training set showed (Figure 4) that the substages of the TNM system were well differentiated (P < 0.05). However, the survival curve of the testing set showed that the curve of stage III was close to that of stage IVA (P = 0.335). Figure 5 shows the survival analysis of the pTNM staging. In the training set, the survival curve discrimination between stages IA and IB was poor (P = 0.240), and there was no significant difference between stages IIA and IIB and stages IIB and IIIA (P < 0.05). In the testing set, there was no significant difference in survival curves between stages IA and IB, IIA and IIB, and IIIA and IIIB (P > 0.05).
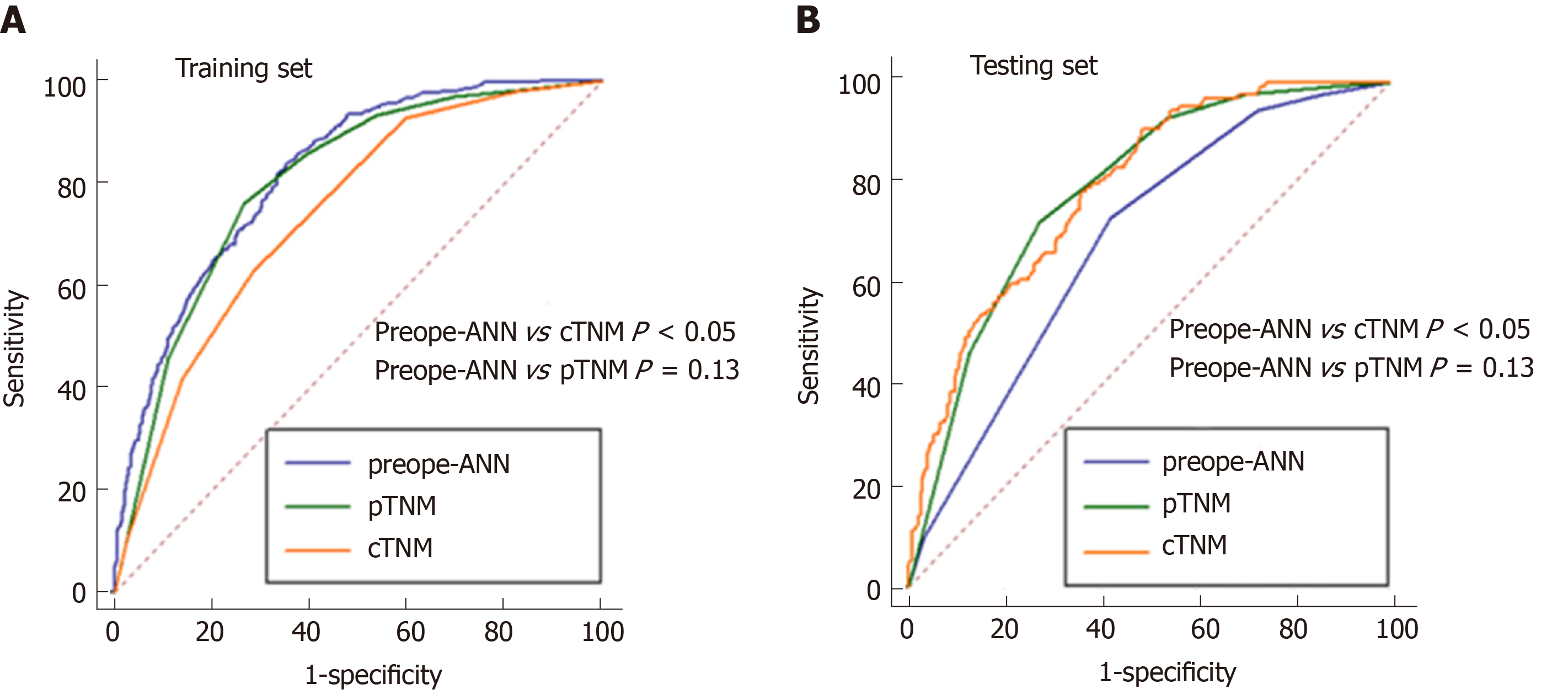
Figure 6 shows the ROC curves of the preope-ANN model, cTNM stage, and pTNM stage in both the training set and the testing set. In the training set, the AUC values of the preope-ANN model, cTNM stage, and pTNM stage were 0.820 (0.800-0.838), 0.740 (0.718-0.762), and 0.803 (0.782-0.822), respectively. The predictive performance of the preope-ANN was better than that of the cTNM stage (P < 0.05) and was similar to that of the pTNM stage (P = 0.130). In the testing set, the AUC values of the preope-ANN model, cTNM stage, and pTNM stage were 0.790 (0.752- 0.825), 0.687 (0.644-0.727), and 0.786 (0.748-0.821), respectively. Comparison of the AUC showed that the prediction performance of the preope-ANN was better than that of the cTNM stage (P < 0.05), and was similar to that of the pTNM (P = 0.858).
As shown in Table 3, for Harrell's C index, the preope-ANN model was superior to the cTNM staging model and had the same performance as the pTNM staging model in both the training and testing sets. (In the training set: Preope-ANN vs cTNM = 0.773 vs 0.663, respectively, P < 0.001; preope-ANN vs pTNM = 0.773 vs 0.757, respectively, P = 0.120; in the testing set: Preope-ANN vs cTNM = 0.752 vs 0.652, respectively, P < 0.001; preope-ANN vs pTNM =0.752 vs 0.740, respectively, P = 0.539). The AIC analysis showed that the preope-ANN model of the training set had a better fitting degree than both the cTNM staging and pTNM staging (preope-ANN vs cTNM = 4977.83 vs 5176.70, respectively, relative likelihood < 0.001; preope-ANN vs pTNM = 4977.83 vs 4999.80, respectively, relative likelihood < 0.001). The fitting degree of the preope-ANN model in the testing set was better than that of the cTNM staging (preope-ANN vs cTNM = 1952.94 vs 2020.37, respectively, relative likelihood < 0.001), and the fitting degree of the preope-ANN model was not inferior to that of the pTNM staging (preope-ANN 1952.94 vs pTNM 1951.84, respectively, relative likelihood = 1.733). Therefore, the performance of the preope-ANN was better than that of the cTNM staging and was similar to that of the pTNM staging.
| Training set | Testing set | Bio-ANN | Cli-ANN | |||||
| Preope-ANN | cTNM | pTNM | Preope-ANN | cTNM | pTNM | |||
| Harrell’s C index | 0.773 (0.753-0.795) | 0.663 (0.640-0.687) | 0.757 (0.735-0.779) | 0.752 (0.719-0.785) | 0.652 (0.615-0.688) | 0.740 (0.707-0.775) | 0.722 (0.698-0.746) | 0.760 (0.738-0.782) |
| P value | < 0.001 | 0.120 | < 0.001 | 0.539 | aP < 0.001; bP = 0.000; cP = 0.018 | dP < 0.001 eP < 0.000; fP = 0.827 | ||
| AIC | 4977.83 | 5176.70 | 4999.80 | 1952.94 | 2020.37 | 1951.84 | 5115.9 | 5011.9 |
| Relative likelihood | < 0.001 | < 0.001 | <0.001 | 1.733 | aP < 0.001; bP > 1 cP < 0.001 | dP = 0.001 E > 1 fP = 0.06 | ||
Presently, GC remains a common malignant tumor worldwide and the third leading cancer cause of death. As is known, it is very important to develop a GC prognostic model in order to provide valuable prognosis information for the patients and help clinicians formulate reasonable treatment regimens for the patients[19]. The TNM staging system proposed by the AJCC is the most important prognostic evaluation system for GC, and it has served as the main instruction for clinicians to choose the treatment plan. However, pTNM staging needs both grouping information and prognostic information from the postoperative histopathology results of the tumor specimen, which prevents the approach from guiding the preoperative treatment decision[24]. The exact pretreatment clinical stage is essential to customize the treatment strategy for each patient. Park et al[25] suggested that the clinical staging based on endoscopy and the CT scan has predictive value, where cTNM staging can be used to guide the treatment of GC patients; however, its accuracy depends on the imaging experience of the physician. Some scholars found that this method had limitations in evaluating the cT stage of large tumors, while the accuracy of cN was only 20%; and over 80% of pN0 patients are overestimated[26,27]. Thus, there is an urgent need for a more accurate preoperative prognostic model to guide the choice of treatment options.
In recent years, increasingly more studies have shown that blood inflammatory markers are associated with a poor prognosis in cancer patients[28]. The NLR, PLR, and PNI in cancer patients have been proven to be prognostic markers for various malignant tumors[29-32]. In our study, logistic analysis confirmed that the preoperative NLR, PLR, PNI, and AGR were significant prognostic factors for the 3-year survival, which is consistent with previous studies. However, because of the nonlinearity of biological information in the human body, the traditional model inevitably has had some limitations when the traditional linear analysis method was used to construct the prognostic model in previous studies. At the same time, the growth of a tumor is a process of interaction between the human body and the tumor, which depends on the nutritional status of the body, the immune system, and the tumor malignancy; thus, the application of a single index for forecasting may lack accuracy[18]. Consequently, we need a new statistical model that can synthesize the biological indicators and better address the nonlinear relationship among the indicators.
The ANN is a new computational model developed by simulating the function of human brain; this method can establish a nonlinear statistical model to evaluate complex biological systems and address the relationship between complex biological indicators more flexibly[33]. In recent years, ANNs have been successfully applied to the field for the identification of lesions in pathological specimens, automatic detection of breast X-ray injury, and disease diagnosis and treatment[33,34].
We synthesized preoperative blood biomarkers (the inflammatory indicators and PNI) and preoperative clinical data to establish the preope-ANN, which are easily available compared with the need for postoperative pathological results. At the same time, we used the ANN to reduce the error of human interference, ensuring the objectivity and accuracy of the results. The verification results showed that the accuracy of the preope-ANN model in predicting the 3-year survival rate was 91.7%. In addition, the comparison of Harrell's C index and AIC analysis showed that the accuracy and the fitting degree of the preope-ANN model were better than those of cTNM staging, and the preope-ANN model could achieve the same prediction effect as pTNM staging. The TNM staging system divides the patients into different risk groups, and our preope-ANN model can provide an even more detailed prediction for each patient, which is better than grouping the predictions. The preope-ANN model can be used to predict the long-term survival of patients before surgery and to choose a reasonable individualized treatment according to the prognosis. We can obtain the possible poor prognosis information of those patients with a low score before surgery and improve the prognosis by adopting neoadjuvant radiotherapy and chemotherapy.
This study still has some limitations. First, some patients had a follow-up period less than 5 years, and we only conducted the study for the 3-year survival outcome, not for the longer-term survival outcome. Second, this was a retrospective study, and some potential biases were still unavoidable. Moreover, this study included all postoperative patients, and the results are not suitable for the evaluation of the prognosis of patients with unresectable advanced GC. Nevertheless, this study first confirmed that the preope-ANN is a novel and convenient prognostic model through the use of a large sample data size, which can effectively predict the prognosis of GC patients. In the clinic, the preope-ANN model can be considered as part of preoperative risk stratification to guide the individualized treatment of patients with GC. The next challenge is to establish a web version of the preope-ANN model that can be dynamically adjusted for the input of different sample data; with this approach, the model accuracy would be closer to the real value and more flexibly applied to the evaluation of clinical patients.
Because of the powerful abilities of self-learning and handling complex biological information, artificial neural network (ANN) models have been widely applied to disease diagnosis, imaging analysis, and prognosis prediction. However, there has been no trained preoperative ANN (preope-ANN) model to preoperatively predict the prognosis of patients with gastric cancer (GC).
This study combined the preoperative blood biomarkers and preoperative tumor data to establish an ANN model in order to build a reliable preoperative prediction system that can achieve the same effect as postoperative TNM staging. The aim of this study was to evaluate the prognosis of patients with GC and to provide a reasonable individualized treatment plan for patients.
We aimed to establish a neural network model that can predict long-term survival of GC patients before surgery to evaluate the tumor condition before the operation.
The clinicopathological data of 1608 GC patients treated from January 2011 to April 2015 at the Department of Gastric Surgery, Fujian Medical University Union Hospital were analyzed retrospectively. Patients were randomly divided into a training set (70%) for establishing a preope-ANN model and a testing set (30%). The prognostic evaluation ability of the preope-ANN model was compared with that of the American Joint Commission on Cancer (8th edition) clinical TNM stage (cTNM) and pathological TNM stage (pTNM) through the receiver operating characteristic curve, Akaike information criterion index, Harrell's C index, and likelihood ratio chi-square.
We used the variables that were statistically significant factors for the 3-year overall survival as input-layer variables to develop a preope-ANN in the training set. The survival curves within each score of the preope-ANN had good discrimination (P < 0.05). Comparing the preope-ANN model, cTNM, and pTNM in both the training and testing sets, the preope-ANN model was superior to cTNM in predictive discrimination (C index), predictive homogeneity (likelihood ratio chi-square), and prediction accuracy (area under the curve). The prediction efficiency of the preope-ANN model was similar to that of pTNM.
The preope-ANN model can accurately predict the long-term survival of GC patients, and its predictive efficiency is not inferior to pTNM staging.
This study for the first time confirmed that the preope-ANN is a novel and convenient prognostic model through the use of a large sample data size, which can effectively predict the prognosis of GC patients. In the clinic, preope-ANN can be considered as part of preoperative risk stratification to guide the individualized treatment of patients with GC. The next challenge is to establish a web version of the preope-ANN model that can be dynamically adjusted for the input of different sample data; with this approach, the model accuracy would be closer to the real value and more flexibly applied to the evaluation of clinical patients.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lazăr DC, de Melo FF, Nagem RG S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50811] [Article Influence: 8468.5] [Reference Citation Analysis (44)] |
| 2. | Dassen AE, Dikken JL, van de Velde CJ, Wouters MW, Bosscha K, Lemmens VE. Changes in treatment patterns and their influence on long-term survival in patients with stages I-III gastric cancer in The Netherlands. Int J Cancer. 2013;133:1859-1866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42955] [Article Influence: 3304.2] [Reference Citation Analysis (4)] |
| 4. | Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543-2546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1890] [Cited by in F6Publishing: 1860] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 5. | Hirahara N, Tajima Y, Fujii Y, Yamamoto T, Hyakudomi R, Taniura T, Kaji S, Kawabata Y. Preoperative Prognostic Nutritional Index Predicts Long-term Outcome in Gastric Cancer: A Propensity Score-matched Analysis. Anticancer Res. 2018;38:4735-4746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5245] [Cited by in F6Publishing: 5418] [Article Influence: 235.6] [Reference Citation Analysis (0)] |
| 7. | Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10123] [Cited by in F6Publishing: 10557] [Article Influence: 479.9] [Reference Citation Analysis (0)] |
| 8. | Roxburgh CS, McMillan DC. Role of systemic inflammatory response in predicting survival in patients with primary operable cancer. Future Oncol. 2010;6:149-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 607] [Cited by in F6Publishing: 692] [Article Influence: 49.4] [Reference Citation Analysis (1)] |
| 9. | Deng Q, He B, Liu X, Yue J, Ying H, Pan Y, Sun H, Chen J, Wang F, Gao T, Zhang L, Wang S. Prognostic value of pre-operative inflammatory response biomarkers in gastric cancer patients and the construction of a predictive model. J Transl Med. 2015;13:66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 10. | Pan QX, Su ZJ, Zhang JH, Wang CR, Ke SY. A comparison of the prognostic value of preoperative inflammation-based scores and TNM stage in patients with gastric cancer. Onco Targets Ther. 2015;8:1375-1385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 11. | Liu X, Wu Z, Lin E, Li W, Chen Y, Sun X, Zhou Z. Systemic prognostic score and nomogram based on inflammatory, nutritional and tumor markers predict cancer-specific survival in stage II-III gastric cancer patients with adjuvant chemotherapy. Clin Nutr. 2019;38:1853-1860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 12. | Cucchetti A, Piscaglia F, Grigioni AD, Ravaioli M, Cescon M, Zanello M, Grazi GL, Golfieri R, Grigioni WF, Pinna AD. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol. 2010;52:880-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 13. | McQuade JL, Daniel CR, Hess KR, Mak C, Wang DY, Rai RR, Park JJ, Haydu LE, Spencer C, Wongchenko M, Lane S, Lee DY, Kaper M, McKean M, Beckermann KE, Rubinstein SM, Rooney I, Musib L, Budha N, Hsu J, Nowicki TS, Avila A, Haas T, Puligandla M, Lee S, Fang S, Wargo JA, Gershenwald JE, Lee JE, Hwu P, Chapman PB, Sosman JA, Schadendorf D, Grob JJ, Flaherty KT, Walker D, Yan Y, McKenna E, Legos JJ, Carlino MS, Ribas A, Kirkwood JM, Long GV, Johnson DB, Menzies AM, Davies MA. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol. 2018;19:310-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 438] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 14. | Liu X, Sun X, Liu J, Kong P, Chen S, Zhan Y, Xu D. Preoperative C-Reactive Protein/Albumin Ratio Predicts Prognosis of Patients after Curative Resection for Gastric Cancer. Transl Oncol. 2015;8:339-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 15. | Ma M, Wang J, Hu Y, Weng M, Liu X, Wang Y. Prognostic Value of Inflammatory Biomarkers in Gastric Cancer Patients and the Construction of a Predictive Model. Dig Surg. 2019;36:433-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Burke HB, Goodman PH, Rosen DB, Henson DE, Weinstein JN, Harrell FE, Marks JR, Winchester DP, Bostwick DG. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer. 1997;79:857-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 17. | Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics--application to complex microarray and mass spectrometry datasets in cancer studies. Brief Bioinform. 2009;10:315-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 18. | Davies AR, Gossage JA, Zylstra J, Mattsson F, Lagergren J, Maisey N, Smyth EC, Cunningham D, Allum WH, Mason RC. Tumor stage after neoadjuvant chemotherapy determines survival after surgery for adenocarcinoma of the esophagus and esophagogastric junction. J Clin Oncol. 2014;32:2983-2990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 19. | Marrelli D, Morgagni P, de Manzoni G, Coniglio A, Marchet A, Saragoni L, Tiberio G, Roviello F; Italian Research Group for Gastric Cancer (IRGGC). Prognostic value of the 7th AJCC/UICC TNM classification of noncardia gastric cancer: analysis of a large series from specialized Western centers. Ann Surg. 2012;255:486-491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 20. | Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 63] [Reference Citation Analysis (0)] |
| 21. | Yoon HM, Ryu KW, Nam BH, Cho SJ, Park SR, Lee JY, Lee JH, Kook MC, Choi IJ, Kim YW. Is the new seventh AJCC/UICC staging system appropriate for patients with gastric cancer? J Am Coll Surg. 2012;214:88-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 22. | Vrieze SI. Model selection and psychological theory: a discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychol Methods. 2012;17:228-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 705] [Cited by in F6Publishing: 691] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 23. | Edeline J, Blanc JF, Johnson P, Campillo-Gimenez B, Ross P, Ma YT, King J, Hubner RA, Sumpter K, Darby S, Evans J, Iwuji C, Swinson D, Collins P, Patel K, Muazzam I, Palmer DH, Meyer T. A multicentre comparison between Child Pugh and Albumin-Bilirubin scores in patients treated with sorafenib for Hepatocellular Carcinoma. Liver Int. 2016;36:1821-1828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 24. | Krittanawong C, Johnson KW, Rosenson RS, Wang Z, Aydar M, Baber U, Min JK, Tang WHW, Halperin JL, Narayan SM. Deep learning for cardiovascular medicine: a practical primer. Eur Heart J. 2019;40:2058-2073. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 159] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 25. | Park SR, Kim MJ, Ryu KW, Lee JH, Lee JS, Nam BH, Choi IJ, Kim YW. Prognostic value of preoperative clinical staging assessed by computed tomography in resectable gastric cancer patients: a viewpoint in the era of preoperative treatment. Ann Surg. 2010;251:428-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Shimizu K, Ito K, Matsunaga N, Shimizu A, Kawakami Y. Diagnosis of gastric cancer with MDCT using the water-filling method and multiplanar reconstruction: CT-histologic correlation. AJR Am J Roentgenol. 2005;185:1152-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 84] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Blank S, Bläker H, Schaible A, Lordick F, Grenacher L, Buechler M, Ott K. Impact of pretherapeutic routine clinical staging for the individualization of treatment in gastric cancer patients. Langenbecks Arch Surg. 2012;397:45-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Zheng L, Zou K, Yang C, Chen F, Guo T, Xiong B. Inflammation-based indexes and clinicopathologic features are strong predictive values of preoperative circulating tumor cell detection in gastric cancer patients. Clin Transl Oncol. 2017;19:1125-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Shimada H, Takiguchi N, Kainuma O, Soda H, Ikeda A, Cho A, Miyazaki A, Gunji H, Yamamoto H, Nagata M. High preoperative neutrophil-lymphocyte ratio predicts poor survival in patients with gastric cancer. Gastric Cancer. 2010;13:170-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 286] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Wang Y, Zhang X, Zhang T. Pretreatment prognostic nutritional index as a prognostic factor in lung cancer: Review and meta-analysis. Clin Chim Acta. 2018;486:303-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 31. | Mohri Y, Inoue Y, Tanaka K, Hiro J, Uchida K, Kusunoki M. Prognostic nutritional index predicts postoperative outcome in colorectal cancer. World J Surg. 2013;37:2688-2692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 161] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 32. | Eo WK, Chang HJ, Suh J, Ahn J, Shin J, Hur JY, Kim GY, Lee S, Park S, Lee S. The Prognostic Nutritional Index Predicts Survival and Identifies Aggressiveness of Gastric Cancer. Nutr Cancer. 2015;67:1260-1267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Kim JH, Kim HS, Seo WY, Nam CM, Kim KY, Jeung HC, Lai JF, Chung HC, Noh SH, Rha SY. External validation of nomogram for the prediction of recurrence after curative resection in early gastric cancer. Ann Oncol. 2012;23:361-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 2018;21:249-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |