Manuscript accepted on :March 09, 2018
Published online on: --
Plagiarism Check: Yes
Sri Masyeni , Hegard Sukmawati, Ayu Savitri Siskayani, Satya Dharmayanti and Kartika Sari
, Hegard Sukmawati, Ayu Savitri Siskayani, Satya Dharmayanti and Kartika Sari
Faculty of Medicine and Health Science, University of Warmadewa, Jln Terompong 24, Denpasar-Bali, Indonesia 80235.
Corresponding Author E-mail: masyeniputu@yahoo.com
DOI : https://dx.doi.org/10.13005/bpj/1399
Abstract
Antimicrobial resistance (AMR) is emerging global health problem worldwide. Resistant bacteria generate higher morbidity and mortality rates. Lack of awareness of AMR includes self-antibiotic prescription, lack of access to get the bacteria and antibiograms data were leading factors for AMR development. The objective of the study was to determine the profile of bacteria and antimicrobial susceptibility patterns of different specimens among two referred microbial laboratories in Denpasar Bali. A retrospective data from January 2015 to December 2016 of various specimens in two different laboratories were reviewed. Type of clinical specimen, type of bacterial isolate and antimicrobial susceptibility pattern from different isolates were extracted using data extraction format. of the 760 various specimens analysed, pathogens were identified in 717 (94.3%) specimens. Almost all of the specimens indicated more than 90% positive cultured result. In contrast with the blood specimens which detected only 50% pathogens. The big five bacteria found were Staphylococcus spp, Escherichia coli, Streptococcus spp, Enterobacter spp and Pseudomonas aeruginosa. These five bacteria were found to have sensitivity rate more than 60% to gentamycin, around 50% to ciprofloxacin, and very low sensitivity to erythromycin (0-15%). Of 63 Pseudomonas aeruginosa isolates, 97% exhibited resistance to erythromycin, and 84%, 83% resistance to cefuroxime and amoxicillin, respectively. Similar resistance pattern also showed by Escherichia coli whereas 100% of these pathogens resistance to erythromycin, followed by 83% resistance to amoxicillin and 81% resistance to cefuroxime. The highest multidrug-resistance rate was observed in Staphylococcus spp isolates (62%), in reverse with only 17% MDR of Proteus sp. The five predominant bacteria isolates showed high resistance to erythromycin. Multidrug-resistant was common in the present study in which Pseudomonas aeruginosa and Staphylococcus spp identified as the most multi drugs resistant pathogens. Gentamycin was the most effective antibiotic against most of the bacteria. Periodic surveillance to determine the pattern of bacteria and antibiotic sensitivity is recommended for generating a local antibiograms for physician guidelines in combating an infection.
Keywords
Antimicrobial; Bacteria; Susceptibility; Specimens Various;
Download this article as:| Copy the following to cite this article: Masyeni S, Sukmawati H, Siskayani A. S, Dharmayanti S, Sari K. Antimicrobial Susceptibility Pattern of Pathogens Isolated from Various Specimens in Denpasar-Bali: A Two Years Retrospective Study. Biomed Pharmacol J 2018;11(1). |
| Copy the following to cite this URL: Masyeni S, Sukmawati H, Siskayani A. S, Dharmayanti S, Sari K. Antimicrobial Susceptibility Pattern of Pathogens Isolated from Various Specimens in Denpasar-Bali: A Two Years Retrospective Study. Biomed Pharmacol J 2018;11(1). Available from: http://biomedpharmajournal.org/?p=19401 |
Introduction
Antimicrobial resistant is the threat rising star to the people particularly in the most poverty countries.1,2 Factors contributed to these are antibiotics misuse or widespread use by the health professional, poor drug quality, high incidence of infections, unhygienic condition and lack of AMR surveillance.2,3 Infections caused by resistant pathogens confer high morbidity and mortality rate by reducing the efficacy of an antibiotic, antiparasitic, antifungal or antiviral drug.4-7 Publication of AMR particularly in antibacterial resistance (ABR) has been surprisingly increasing in several decades related to very high rate resistance of bacteria has been observed.8-11 One to others studies result refers that the pattern of the organism and resistance was changed over time. Even from one geographical area, the organism and AMR profile difference time to time, specimens to specimens. Escherichia coli was the most (66.7%) bacterial isolated with Extended-Spectrum Beta-Lactamase (ESBL) resistance seen from the urinary specimen in South India.12 In contrast with other studies in India and Bangladesh, this found Enterococci as the most bacteria isolated from patients with urinary tract infection.13,14 Different specimens revealed different microorganisms, Acinetobacter baumannii reported as the most common cause of a ventilator-associated pneumonia and confers mortality rate as high as 41.4%15 Coagulase-negative Staphylococci found in septicaemic burn patients.16 Enterobacteriaceae was fou, nd as high as 73.2% in diabetic foot infection and with the increasing of Wagener’s grade, the proportion of gram-negative bacterial infection particularly Pseudomonas was an increase.17 Staphylococcus aureus was recovered as high as 14,8(194/1360)% from different isolate but mainly on the pus/abscess isolate, in which Methicillin-Resistant Staphylococcus aureus (MRSA) found out 17.4%.18
Bactericidal antibiotics induced bacterial cell death by inhibiting synthesis of bacterial cell wall, DNA or RNA, proteins, competitive inhibition of folic acid or act as membrane disorganizing agents.19,20 Mechanism of resistance can be natural/intrinsic or extrinsic that transmitted vertically or horizontally.21 The bacteria may resistant to one or more antibiotics. Multiple-drug resistance defines as resistance to ≥1 agent in three or more class or antibiotic category, and the term of Extensively drugs resistant (XDR) used for organisms resistant to ≥1 agent in ≤ 2 class or antibiotic category. 22 Isolates with MDR were documented for 40.5% from different grades of diabetic foot infection and XDR accounted for 9.7% of bacteria.17 High mortality rate accounted for 76.9% of septic burn patient documented in Jakarta, related to MDR Pseudomonas aeruginosa (33.3%) and Klebsiella pneumonia (28.9%).23 The high impact of the resistant microorganism and temporal changes of bacterial isolates from time to time reveal that it is very important to report regularly the bacterial and susceptibility testing of the specimens worldwide to guide the physician to choose the appropriate antibiotic in the management of bacterial infection. The current study aim was to define the pattern of bacterial and susceptibility test result from two referred laboratories in Denpasar-Bali.
Material and Methods
Retrospective studies on the data of culture report were collected from two laboratories during 2015 through 2016. The Quantum and Bali Province Laboratories in Denpasar-Bali, are the two referred laboratories who received specimens not only from Denpasar but also others municipalities in Bali. A total of 767 various specimens was annualized from various areas in Bali. Instead type of clinical specimens, we also collected age, sex, type of bacterial isolate, and pattern of antimicrobial susceptibilities were collected using data extraction format.
The culture and identification were done on the specimens according to the Standard Operation Procedure of the Microbiology Department of the laboratories. The blood MacConkey and Chocolate Agar were the culture media used to isolated microorganism on the specimens. After adjustment to 0.5 McFarland, a standard inoculum was swabbed on Muller Hinton agar and accompanied with immersing for 2-5 minutes. The antibiotic disc were conceived and pressed on the media, incubated at 370C for 24 hours. Identification of the microorganism was based on the morphology of the colony and biochemical tests. Antimicrobial resistance testing was carried out by using the Kirby-Bauer disc diffusion methods and was reported in conformity with Clinical Laboratory Standard Institute (CLSI) guideline. The invitro antibiotic testing towards the antibiotics such as amoxicillin (30 µg), ampicillin (10 µg), chloramphenicol (30 µg), kanamycin (30 µg), nitrofurantoin (300), nalidixic acid (30 µg), tetracyclin (30 µg), cephazolin (30 µg), trimethoprim (5 µg), norfloxacin (10 µg), amikacin (10 µg), erythromycin (15 µg), cefuroxime (30 µg), streptomycin (10 µg), neomycin (30 µg), ceftazidime (30 µg), ciprofloxacin (3 µg), gentamycin (10 µg), cefotaxime (30 µg), ceftriaxone (30 µg).
Drug Resistance
The classification of the drug resistance as below:
One drug resistance is resistant to one class of antibiotic.
Multidrug resistance is resistant to ≥1 agent in three or more class or antibiotic category,
Extensively drugs resistant (XDR) is resistant to ≥1 agent but two or fewer antimicrobial categories (22).
Data Analysis
Data was analysed with excel and presented descriptively
Ethics statement
This study was conducted with approval from the Medical Research Ethics Committees of Faculty of Medicine, Udayana University (document number 145/UN.14.2/KEP/2017).
Result
A total of 760 specimens were received in the two laboratories during 2015 to 2016, in which 717 (94.3%) pathogens were detected. Urine 175 (23.0%), pus 164 (21.6%), and sputum 108 (14.2%) were the most frequent samples processed. High positive rate (93 to 100%) of microbial isolation observed from all the specimen types, except for blood specimens (50%). Gram negative bacteria were more dominant bacteria found than Gram positive bacteria in the total specimens (68.6% vs 31.4%). The most frequent Gram negative bacteria found was Escherichia coli (21%), and the Gram positive bacteria was Staphylococcus spp. (32%). Overall, the big five isolates found were Staphylococcus spp (32%), Escherichia coli (21%), Streptococcus spp (13%), Enterobacter sp (10%), and Pseudomonas aeruginosa (9%).
Staphylococcus spp was the most pathogens found on the blood, skin scratches, and pus specimens, meanwhile Escherichia coli was the major pathogens found on the urine and faeces specimens. Streptococcus spp mostly found on the sputum specimens. The more details pathogens pattern in the current study are presented on Table 1.
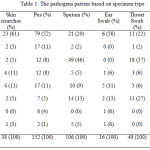 |
Table 1: The pathogens pattern based on specimen type
|
The in vitro antibiotic resistance test of gram negative bacteria were ranged between 12 to 100%. Escherichia coli isolates were 100% resistance to erythromycin, 83% to amoxicillin, and 81% to cefuroxime. Likewise, Enterobacter sp. were resistance to erythromycin (100%), (75%) to amoxicillin, and cefuroxime (65%). Isolates of Pseudomonas aeruginosa also displayed resistance to erythromycin (97%), amoxicillin (83%) and cefuroxime (84%) with addition of resistance to chloramphenicol (75%) and cefotaxime (66%). On the other hand, lower resistance was seen to gentamycin antibiotic (table 2).
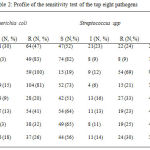 |
Table 2: Profile of the sensitivity test of the top eight pathogens
|
The percentage resistance of Gram positive isolates were ranged between 9 to 83%. Staphylococcus spp showed high resistance to erythromycin (83%), ceftazidime (68%), and amoxicillin (62%). Streptococcus spp revealed high resistance to erythromycin (69%) but not to the other antibiotics. Both types of Gram positive isolates showed quite high sensitivity to Gentamicin which were 62% in Staphylococcus spp and 65% in Streptococcus spp. (table 2).
Overall, the study revealed high proportion of resistance of Staphylococcus spp, Escherichia coli, Streptococcus spp, Enterobacter sp, and Pseudomonas aeruginosa towards erythromycin, with magnitude ranged between 69% and 100%. However, they were still susceptible to gentamycin, in which each bacteria shown sensitivity rate of more than 50%.
Table 3: Multidrug Resistance Profile
| Isolates (N) | MDR (N, %) |
| Staphylococcus spp (221) | 137 (62%) |
| P. aeruginosa (59) | 33 (56%) |
| Enterobacter sp (67) | 35 (52%) |
| Escherichia coli (144) | 70 (49%) |
| Klebsiella sp (68) | 33 (49%) |
| Streptococcus spp (96) | 27 (28%) |
| Proteus spp (12) | 2 (17%) |
Data comparison of 2015 and 2016 showed an increase of antibiotic resistance of Staphylococcus spp and Streptococcus spp to chloramphenicol, amoxicillin, and ciprofloxacin. Similarly, Escherichia coli resistance increased to the three antibiotics, but not to amoxicillin. The resistance rate of Escherichia coli to amoxicillin was 52% in 2015 and decreased to 12% in 2016. While, Enterobacter sp, and Pseudomonas aeruginosa showed an increase resistance to chloramphenicol. Figure 1 indicates that Streptococcus spp revealed relatively lower resistance to all classes of antibiotic year to year than the other isolates.
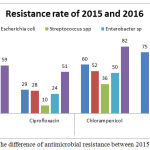 |
Figure 1: The difference of antimicrobial resistance between 2015 and 2016
|
In general, MDR was identified as high as 342/717 (47.7%) of all isolates. The five types of isolate showed resistance to three or more antibiotic classes with various magnitudes. The most apparent multidrug resistance established by Staphylococcus spp and Pseudomonas aeruginosa. This multiple drug resistance found in 62% of Staphylococcus spp, 56% of Pseudomonas aeruginosa, 49% of Escherichia coli, 52% of Enterobacter sp, and 28% of Streptococcus spp. Dissecting into specimen types, MDR mostly found in urine specimen (26.9%) followed with pus (21.6%), and faeces specimen (11.4%). Escherichia coli was the most MDR bacteria found in urine and faeces specimens. In contrast with the MDR’s Staphylococcus spp, that mostly found in pus specimen. There was decreasing MDR trend from 30% to 17.7% of the specimens in 2015 to 2016.
Discussion
Infections are major problem in developing countries such as Indonesia whereas the hygiene and sanitation remain below the international standard rule. Lack of microbial and antimicrobial data are also a problem in guiding the physician in treating the patients with an infection before the definitive treatment applied for the best outcome. Time to time the pattern of infecting microorganism always changes and needs regular investigations in order to provide the update data whether the microorganism itself and also the profile of antimicrobial resistance. Several studies about antimicrobial resistance from Indonesia have been reported from diarrheal patients 24 various specimens,11,25 burn patients 23 and urine.26 More specific specimens reported from ear pus discharge,27 prosthetic joint infection,28 bloodstream infection.29,30
This study revealed that urine, pus, and sputum were the most frequent samples processed and they also showed high positive rate (93 to 100%) of microbial isolation. While, blood sample was the only specimen with low proportion of positive culture. In contrast to our study, Moolchandani, Sastry 31 found the tracheal aspirate as the most specimens with positive culture in the intensive care unit, followed by exudate, urine and blood specimens. Different study settings and geographic could be the reason for the difference in the finding.
Gram negative bacteria were found more frequent than Gram positive bacteria in the total specimens (68.6% vs 31.4%). Similar reports were published by Nurmala, Virgiandhy,32 Setiawan 33 and Sianturi, Hasibuan.34 These three studies were conducted in hospital setting in Indonesia and the results indicated that Gram negative bacterial infections are more common in Indonesia.
The main bacteria isolated from the urine specimens was Escherichia coli. This result was in line with study by Bitew, Molalign 35 which also found Escherichia coli as the dominant bacterium in urine samples and has the least susceptibility to erythromycin. Another study found Escherichia coli producing Extended Spectrum Beta-Lactamase (ESBL) enzyme. The ESBL Escherichia coli confers resistance to the third generation of cephalosporin such as cefotaxime and ceftazidime.26 In contrast with other study result that found Enterococci from the urine specimen and resistant to more than three class of antibiotics such as amoxicillin, co-trimoxazole, ciprofloxacin, gentamycin, ceftriaxone and cefuroxime. 13 Overall, Escherichia coli in the current study possess high resistant all class of antibiotics particularly to erythromycin (100%), amoxicillin (83%) and cefuroxime (80.6%). Escherichia coli producing ESBL found not only in urine specimens but also in various specimens. 11 Escherichia coli also found in febrile patient with septic syndrome.
The main bacteria found in sputum were Streptococcus spp that still showed moderate sensitivity to many antibiotics. Its sensitivity rate was more than 50% to amoxicillin, ciprofloxacin, cefotaxime, gentamycin, ceftazidime, cefuroxime, but only 19.2% to erythromycin. Another study was successfully isolate anaerobes bacteria primarily the genera of Prevotella, Veillonella, Propionibacterium and Actinomyces from sputum of the patients with cystic fibrosis.36 The difference study results may be due to different time and site of the studies.
Blood stream infections are associated with high morbidity and mortality elsewhere. 37 The top bacteria isolated from the blood specimen was Staphylococcus spp, that possess low sensitivity to chloramphenicol (29.6%), amoxicillin (27.9%), erythromycin (7.1%), ceftazidime (16.8%). Methicillin resistant Staphylococcus aureus was isolated from blood specimens of septic pediatric patients.38 Another study found Staphylococcus aureus in febrile patients.30
Gram negative isolates were highly resistant to erythromycin, amoxicillin, and cefuroxime that differ with other study finding in Iran.39 The study by 40 found that the most Gram- negative bacilli were Escherichia coli, Klebsiella pneumonia and Pseudomonas aeruginosa that highly resistance to the third generation of cephalosporin. The contrary data possibly caused by different study setting and different prescription pattern of antibiotic in both countries. Self-antibiotic prescription is quite common behaviour in Bali. However, there is no official report to support this suspicion.
Overall, the isolates showed high resistance to erythromycin and quite high sensitivity to gentamicin. Another study on pus samples of Otitis Media patient support the current finding of the sensitivity to gentamycin.41 In general, the use of gentamycin in community setting is very rare since there is no oral dosage form of gentamycin. This fact may explain why the pathogens remain sensitive to gentamycin.
Among the pathogens detected in the current study, Staphylococcus spp and Pseudomonas aeruginosa were the top two pathogens with multidrug resistance, particularly on the pus specimen. They are the member of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens which acts as the leading cause of nosocomial infections worldwide.42 Most of them are MDR bacteria. Staphylococcus aureus, one of the species of Staphylococcus spp, is a common cause of infective endocarditis,43 skin and soft tissues infection, 44 pleuropulmonary infection 45 and others. The methicillin resistance S. aureus (MRSA) is associated with poor clinical outcome in numerous infections include prolonged the hospital stay. 45,46 A similar retrospective study revealed the overall Gram-positive MDR was 84.6%, but only 12% of Staphylococcus aureus was MDR to three different antibiotics.47 Lack of Staphylococcus species data may explained the MDR results discrepancies with the previous study. Patil and Patil (2017) reported of total 55 MDR isolates in ventilator-associated pneumonia, 20% was Pseudomonas aeruginosa, and 16.36% was coagulase positive Staphylococcus aureus.48 Again, the difference study result may be due to unavailability of the current study to specify the Staphylococcus genus.
A record based retrospective study has been done to evaluate the pattern of microorganism and antimicrobial resistance in intensive care unit. The study found Escherichia coli as the predominant microorganism in urine, exudate and sterile fluid specimens. The Gram-negative bacilli found as the most MDR, followed by MRSA as high as 40.6% Moolchandani, Sastry.49
In conclusion, most of pathogen isolates in Denpasar showed high resistance to erythromycin, but were susceptible to gentamycin. Multidrug-resistant was common in which Pseudomonas aeruginosa and Staphylococcus spp identified as the most multi drugs resistant pathogens. The incoherent finding among the current study and other studies above reflects the variability and the dynamic of the microorganism in various areas. These finding suggest continuing and periodic evaluation of microbiological pattern and sensitivity test to provide the update data for clinicians in choosing the appropriate antibiotic for the optimum outcome.
Acknowledgement
We acknowledge Bali Quantum Clinical Laboratory and Bali Province Laboratories for providing the data. We have special thanks to Warmadewa University for the financial support.
References
- Hart C., Kariuki S. Antimicrobial resistance in developing countries. BMJ. British Medical Journal. 1998;317(7159):647.
CrossRef - Organization W. H. Antimicrobial resistance global report on surveillance: World Health Organization. 2014.
- Okeke I. N., Lamikanra A., Edelman R. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging infectious diseases. 1999;5(1):18.
CrossRef - Group W. Y. I. S. Bacterial etiology of serious infections in young infants in developing countries: results of a multicenter study. The Pediatric infectious disease journal. 1999;18(10):S17-S22.
- Hagan P., Blumenthal U. J., Dunn D., Simpson A. J., Wilkins H. A. Human IgE, IgG4 and resistance to reinfection with Schistosomahaematobium. Nature. 1991;349(6306):243-5.
CrossRef - Margolis S., Oie H., Levy H. The effect of interferon, interferon inducers or interferon induced virus resistance on subsequent interferon production. Journal of General Virology. 1972;15(2):119-28.
CrossRef - Sanglard D., Kuchler K., Ischer F., Pagani J., Monod M., Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrobial agents and chemotherapy. 1995;39(11):2378-86.
CrossRef - Duttaroy B., Mehta S. Extended spectrum b lactamases (ESBL) in clinical isolates of Klebsiella pneumoniae and Escherichia coli. Indian J Pathol Microbiol. 2005;48(1):45-8.
CrossRef - Geslin P., Buu-Hoi A., Fremaux A., Acar J. Antimicrobial resistance in Streptococcus pneumoniae an epidemiological survey in France, 1970–1990. Clinical Infectious Diseases. 1992;15(1):95-8.
- Reacher M. H., Shah A., Livermore D. M., Wale M. C., Graham C., Johnson A. P., et al. Bacteraemia and antibiotic resistance of its pathogens reported in England and Wales between 1990 and 1998: trend analysis. Bmj. 2000;320(7229):213-6.
CrossRef - Severin J. A., Mertaniasih N. M., Kuntaman K., Lestari E. S., Purwanta M., Lemmens-Den T. N., et al. Molecular characterization of extended-spectrum β-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya Indonesia. Journal of antimicrobial chemotherapy. 2010;65(3):465-9.
CrossRef - Eshwarappa M., Dosegowda R., Aprameya I. V., Khan M. W., Kumar P. S., Kempegowda P. Clinico-microbiological profile of urinary tract infection in south India. Indian Journal of Nephrology. 2011;21(1):30-6.
CrossRef - Akhter J., Ahmed S., Anwar S. Antimicrobial Susceptibility Patterns of Enterococcus species Isolated from urinary tract infections. Bangladesh Journal of Medical Microbiology. 2017;8(1):16-20.
CrossRef - Goel V., Kumar D., Kumar R., Mathur P., Singh S. Community Acquired Enterococcal Urinary Tract Infections and Antibiotic Resistance Profile in North India. Journal of laboratory physicians. 2016;8(1):50-4.
CrossRef - Chittawatanarat K., Jaipakdee W., Chotirosniramit N., Chandacham K., Jirapongcharoenlap T. Microbiology, resistance patterns and risk factors of mortality in ventilator-associated bacterial pneumonia in a Northern Thai tertiary-care university based general surgical intensive care unit. Infection and Drug Resistance. 2014;7:203-10.
CrossRef - Sewunet T., Demissie Y., Mihret A., Abebe T. Bacterial Profile and Antimicrobial Susceptibility Pattern of Isolates Among Burn Patients at Yekatit 12 Hospital Burn Center, Addis Ababa, Ethiopia. Ethiopian Journal of Health Sciences. 2013;23(3):209-16.
CrossRef - Xie X., Bao Y., Ni L., Liu D., Niu S., Lin H., et al. Bacterial Profile and Antibiotic Resistance in Patients with Diabetic Foot Ulcer in Guangzhou, Southern China Focus on the Differences among Different Wagner’s Grades, IDSA/IWGDF Grades, and Ulcer Types. International Journal of Endocrinology. 2017;2017:8694903.
CrossRef - Dilnessa T., Bitew A. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolated from clinical samples at Yekatit 12 Hospital Medical College, Addis Ababa, Ethiopia. BMC Infect Dis. 2016;16:398.
CrossRef - Alanis A. J. Resistance to antibiotics: are we in the post-antibiotic era? Archives of medical research. 2005;36(6):697-705.
CrossRef - Kohanski M. A., Dwyer D. J., Collins J. J. How antibiotics kill bacteria from targets to networks. Nature reviews Microbiology. 2010;8(6):423.
CrossRef - Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiology and molecular biology reviews . MMBR. 2010;74(3):417-33.
CrossRef - Magiorakos A. P., Srinivasan A., Carey R., Carmeli Y., Falagas M., Giske C., et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection. 2012;18(3):268-81.
CrossRef - Wardhana A., Djan R., Halim Z. Bacterial and antimicrobial susceptibility profile and the prevalence of sepsis among burn patients at the burn unit of Cipto Mangunkusumo Hospital. Annals of burns and fire disasters. 2017;30(2):107-15.
- Tjaniadi P., Lesmana M., Subekti D., Machpud N., Komalarini S., Santoso W., et al. Antimicrobial resistance of bacterial pathogens associated with diarrheal patients in Indonesia. The American journal of tropical medicine and hygiene. 2003;68(6):666-70.
CrossRef - Rostina R., Rusli B., Arief M., Hardjoeno H. Microbial Patterns Based on Type of Spesimens and its Sensitivity to Antimicrobial Drugs. INDONESIAN JOURNAL OF CLINICAL PATHOLOGY AND MEDICAL LABORATORY. 2017;13(1):13-6.
- Warganegara E., Apriliana E. The Determining type of Extended-Spectrum B-Lactamase Enzyme (ESBL) from Escherichia coil resistance Cephalosporine of third Generation in RSUD Abdoel Moeloek Bandar Lampung. Jurnal Kedokteran. 2014;4(01).
- Mohanna B. M. A., Bahannan A. A. Bacterial Profile And Antibiogram Of Otitis Media Among Children In Yemen. Journal of Ayub Medical College, Abbottabad . JAMC. 2016;28(3):480-3.
- Jordan R. W., Smith N. A., Saithna A., Sprowson A. P., Foguet P. Sensitivities, specificities, and predictive values of microbiological culture techniques for the diagnosis of prosthetic joint infection. BioMed research international. 2014;2014:180416.
CrossRef - Gohel K., Jojera A., Soni S., Gang S., Sabnis R., Desai M. Bacteriological Profile and Drug Resistance Patterns of Blood Culture Isolates in a Tertiary Care Nephrourology Teaching Institute. BioMed research international. 2014;2014:153747.
CrossRef - Wasihun A. G., Wlekidan L. N., Gebremariam S. A., Dejene T. A., Welderufael A. L., Haile T. D., et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. SpringerPlus. 2015;4:314.
CrossRef - Moolchandani K., Sastry A. S., Deepashree R., Sistla S., Harish B., Mandal J. Antimicrobial Resistance Surveillance among Intensive Care Units of a Tertiary Care Hospital in Southern India. Journal of clinical and diagnostic research. JCDR. 2017;11(2):DC01.
- Nurmala N., Virgiandhy I., Andriani A., Liana D. F. Resistensi dan Sensitivitas Bakteri terhadap Antibiotik di RSU dr. Soedarso Pontianak Tahun 2011-2013. eJournal Kedokteran Indonesia. 2015.
- Setiawan M. W. Pola Kuman Pasien yang dirawat di Ruang Rawat Intensif RSUP Dr. Kariadi Semarang: Faculty of Medicine. 2010.
- Sianturi P., Hasibuan B. S., Lubis B. M., Azlin E., Tjipta G. D. Gambaran Pola Resistensi Bakteri di Unit Perawatan Neonatus. Sari Pediatri. 2016;13(6):431-6.
CrossRef - Bitew A., Molalign T., Chanie M. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC infectious diseases. 2017;17(1):654.
CrossRef - Tunney M. M., Field T. R., Moriarty T. F., Patrick S., Doering G., Muhlebach M. S., et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. American journal of respiratory and critical care medicine. 2008;177(9):995-1001.
CrossRef - Diekema D., Beekmann S., Chapin K., Morel K., Munson E., Doern G. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. Journal of Clinical Microbiology. 2003;41(8):3655-60.
CrossRef - Parajuli N. P., Parajuli H., Pandit R., Shakya J., Khanal P. R. Evaluating the Trends of Bloodstream Infections among Pediatric and Adult Patients at a Teaching Hospital of Kathmandu, Nepal Role of Drug Resistant Pathogens. The Canadian Journal of Infectious Diseases & Medical Microbiology = Journal Canadien des Maladies Infectieuses et de la Microbiologie Médicale. 2017;2017:8763135.
CrossRef - Mohammadi-Mehr M., Feizabadi M. Antimicrobial resistance pattern of Gram-negative bacilli isolated from patients at ICUs of Army hospitals in Iran. Iranian journal of microbiology. 2011;3(1):26-30.
- Mohammadi-Mehr M., Feizabadi M. Antimicrobial resistance pattern of Gram-negative bacilli isolated from patients at ICUs of Army hospitals in Iran. Iranian journal of microbiology. 2011;3(1):26.
- Basnet R., Sharma S., Rana J. C., Shah P. K. Bacteriological Study of Otitis Media and Its Antibiotic Susceptibility Pattern. Journal of Nepal Health Research Council. 2017;15(2):124-9.
CrossRef - Santajit S., Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed research international. 2016;2016:2475067.
- Fernandez-Hidalgo N., Ribera A., Larrosa M.N., Viedma E., Origuen J., de Alarcon A., et al. Impact of Staphylococcus aureus phenotype and genotype on the clinical characteristics and outcome of infective endocarditis. A multicenter, longitudinal, prospective, observational study. Clinical microbiology and infection the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2017.
- Nurjadi D., Friedrich-Janicke B., Schafer J., Genderen P. V. J., Goorhuis A., Perignon A., et al. Skin and soft tissue infections in intercontinental travellers and the import of multi-resistant Staphylococcus aureus to Europe. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2015;21(6):567.e1-10.
- Liu F., Wen Z., Wei J., Xue H., Chen Y., Gao W., et al. Epidemiology, microbiology and treatment implications in adult patients hospitalized with pneumonia in different regions of China a retrospective study. Journal of thoracic disease. 2017;9(10):3875-87.
CrossRef - Pulido-Cejudo A., Guzman-Gutierrez M., Jalife-Montano A., Ortiz-Covarrubias A., Martinez-Ordaz J. L., Noyola-Villalobos H. F., et al. Management of acute bacterial skin and skin structure infections with a focus on patients at high risk of treatment failure. Therapeutic advances in infectious disease. 2017;4(5):143-61.
CrossRef - Mulu W., Abera B., Yimer M., Hailu T., Ayele H., Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia a cross-sectional study. BMC research notes. 2017;10(1):254.
CrossRef - Patil H. V., Patil V. C. Incidence, bacteriology and clinical outcome of ventilator-associated pneumonia at tertiary care hospital. Journal of natural science biologyand medicine. 2017;8(1):46-55.
CrossRef - Moolchandani K., Sastry A. S., Deepashree R., Sistla S., Harish B. N., Mandal J. Antimicrobial Resistance Surveillance among Intensive Care Units of a Tertiary Care Hospital in Southern India. Journal of clinical and diagnostic research . JCDR. 2017;11(2):Dc01-dc7.







