-
PDF
- Split View
-
Views
-
Cite
Cite
Dmytro P. Yevtushenko, Rafael Romero, Benjamin S. Forward, Robert E. Hancock, William W. Kay, Santosh Misra, Pathogen-induced expression of a cecropin A-melittin antimicrobial peptide gene confers antifungal resistance in transgenic tobacco, Journal of Experimental Botany, Volume 56, Issue 416, June 2005, Pages 1685–1695, https://doi.org/10.1093/jxb/eri165
Close - Share Icon Share
Abstract
Expression of defensive genes from a promoter that is specifically activated in response to pathogen invasion is highly desirable for engineering disease-resistant plants. A plant transformation vector was constructed with transcriptional fusion between the pathogen-responsive win3.12T promoter from poplar and the gene encoding the novel cecropin A-melittin hybrid peptide (CEMA) with strong antimicrobial activity. This promoter–transgene combination was evaluated in transgenic tobacco (Nicotiana tabacum L. cv. Xanthi) for enhanced plant resistance against a highly virulent pathogenic fungus Fusarium solani. Transgene expression in leaves was strongly increased after fungal infection or mechanical wounding, and the accumulation of CEMA transcripts was found to be systemic and positively correlated with the number of transgene insertions. A simple and efficient in vitro regeneration bioassay for preliminary screening of transgenic lines against pathogenic fungi was developed. CEMA had strong antifungal activity in vitro, inhibiting conidia germination at concentrations that were non-toxic to tobacco protoplasts. Most importantly, the expression level of the CEMA peptide in vivo, regulated by the win3.12T promoter, was sufficient to confer resistance against F. solani in transgenic tobacco. The antifungal resistance of plants with high CEMA expression was strong and reproducible. In addition, leaf tissue extracts from transgenic plants significantly reduced the number of fungal colonies arising from germinated conidia. Accumulation of CEMA peptide in transgenic tobacco had no deleterious effect on plant growth and development. This is the first report showing the application of a heterologous pathogen-inducible promoter to direct the expression of an antimicrobial peptide in plants, and the feasibility of this approach to provide disease resistance in tobacco and, possibly, other crops.
Introduction
Phytopathogens are responsible for most crop losses worldwide and have caused devastating famines in human history (Agrios, 1988; Ali and Reddy, 2000). While conventional breeding remains a major contributor to the production of resistant cultivars, it has limitations due to interspecific sexual incompatibility and the lack of a desired gene pool in donor species, not to mention numerous time-consuming back-crossings. Obviously, transformation of plants by transferring a gene of interest is the logical choice to create disease-resistant cultivars without altering valuable traits of otherwise superior genotypes.
Among different strategies to develop disease-resistant plants, transformation with genes encoding so-called antimicrobial peptides (AMPs) is a promising approach to engineer resistance against a wide range of bacteria, fungi, and even viruses and protozoa (Zasloff, 2002). AMPs represent a diverse family of ubiquitous, naturally occurring defensive molecules found in a variety of multicellular organisms, from primitive invertebrates to plants and animals (Hancock and Lehrer, 1998). They are components of the innate defence mechanisms that host organisms have developed to combat pathogens. Currently, over 500 AMPs have been reported and, regardless of their origin, all these peptides are small (usually less than 50 amino acids), polycationic (i.e. contain excess lysine and arginine residues), and membrane active (Nicolas and Mor, 1995; Hancock and Lehrer, 1998). The main advantage of AMPs over other defensive molecules (i.e. antibiotics) is that they target a fundamental difference in the design of the microbial membranes and the membranes of plants and animals, and therefore the emergence of resistant strains of pathogens is less probable compared with conventional antibiotics (Zasloff, 2002). In addition, because the AMPs are small, they can be readily modified through substitutions, chain elongation or deletions of amino acids to improve their efficiency in specific host–pathogen interactions. Both natural and synthetic AMPs have been shown to inhibit bacterial growth (Powell et al., 1995; Zhong et al., 1995) and germination of fungal conidia (Cavallarin et al., 1998; De Lucca et al., 1998) in vitro. This has prompted research on expressing AMPs in vivo (mostly cecropins and synthetic analogues of cecropin, magainin), with encouraging results on engineering either specific or broad-spectrum disease resistance in tobacco (Jaynes et al., 1993; Huang et al., 1997; DeGray et al., 2001), potato (Gao et al., 2000; Osusky et al., 2000), rice (Sharma et al., 2000), banana (Chakrabarti et al., 2003), and hybrid poplar (Mentag et al., 2003). Hovewer, expression of a native cecropin B or a synthetic cecropin analogue in transgenic tobacco and potato showed no enhanced resistance to bacterial infections, presumably due to degradation of the peptides by host proteases present in the intercellular fluid (Hightower et al., 1994; Allefs et al., 1995; Florack et al., 1995).
Most reports on the expression of AMPs in plants showed an increase in host resistance against bacterial infections, whereas the resistance against fungal infections was less successful. Also, the expression of AMPs was usually under the control of a strong CaMV 35S promoter and its derivatives. This results in constitutive transgene expression throughout the plant at a high level that can have a deleterious effect on plant growth. For practical application of plants with AMPs it is desirable to have transgene expression from a promoter that activates only in response to pathogen invasion. In this paper, a combination of a wound-inducible win3.12T poplar promoter, which is strongly induced in response to fungal infection (Yevtushenko et al., 2004), and a defensive cecropin A-melittin chimeric gene (CEMA) to engineer disease resistance in tobacco plants was evaluated. The CEMA gene encodes a small cationic peptide (28 amino acids, preceded by methionine) with strong antimicrobial activity: it contains eight amino acids residues from cecropin A at the N-terminus, followed by a modified melittin sequence (Hancock et al., 1992). Cecropin A is the main antibacterial component of the immune response in insects (Steiner et al., 1981), and was originally isolated from the haemolymph of the giant silkmoth (Hyalophora cecropia). The N-terminal amphipathic α-helix domain of cecropin A was found to exhibit antifungal activity (Cavallarin et al., 1998). Melittin is the major lytic component in the venom of the honeybee (Apis mellifera). Melittin has excellent antibacterial properties; however, its haemolytic activity presumably makes it unsuitable for expression in transgenic plants. The engineered CEMA combined the biologically active portions of cecropin A and melittin: it was designed using the principles of combinatorial peptide chemistry and plant-optimized codon usage to improve the antimicrobial potency of parent peptides without the undesirable cytotoxic effects of melittin, and for better protection against cleavage by proteases. It is reported here that the CEMA peptide has strong antifungal activity in vitro, inhibiting the germination of conidia at concentrations that are several-fold lower than those causing cytotoxicity in tobacco protoplasts. Data are presented on win3.12T-driven CEMA expression in plants in response to wounding and fungal infection. In addition, a simple and efficient in vitro regeneration bioassay was developed for the preliminary screening of transgenic lines against a pathogenic fungus. Most importantly, it is shown that the expression level of the CEMA peptide, regulated by the win3.12T promoter, was sufficient to confer resistance against F. solani in transgenic tobacco.
Materials and methods
Plant material
Tobacco plants (Nicotiana tabacum L. cv. Xanthi) were grown aseptically in Magenta GA7 vessels (Magenta, USA) on MS medium (Murashige and Skoog, 1962) containing 2% sucrose (pH 5.8), in a 16 h light photoperiod (25 μmol m−2 s−1) at 24 °C.
Vector construction
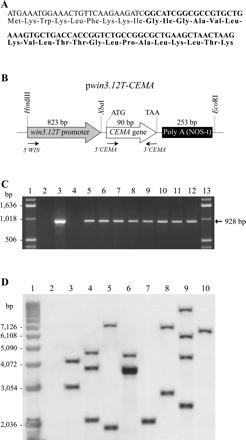
Two binary vectors designed to express genes under the control of a 823 bp pathogen- and wound-inducible promoter win3.12T (Yevtushenko et al., 2004) were used for tobacco transformation: pwin3.12T-CEMA contained the cecropin A-melittin (CEMA) chimeric gene (Hancock et al., 1992), and pwin3.12T-GUS contained the reporter β-D-glucuronidase (GUS) gene (Jefferson et al., 1987). The details on construction of the pwin3.12T-GUS vector are given elsewhere (Yevtushenko et al., 2004). To make pwin3.12T-CEMA, the 363 bp XbaI/EcoRI DNA fragment containing the CEMA gene was ligated into the corresponding sites of pwin3.12T-GUS in place of the deleted GUS-containing region. The result was a transcriptional fusion between the win3.12T promoter, the CEMA coding region and the nopaline synthase 3′-untranslated region. The correct insertion and full nucleotide sequence of the win3.12T promoter and the CEMA gene were confirmed by DNA sequence analysis (PE Aplied Biosystems, USA).
The T-DNA regions of both vectors also contained the NOS-driven NPTII gene, used for the selection of kanamycin-resistant plants. The constructs were introduced into Agrobacterium tumefaciens MP90 by the freeze–thaw method (Holsters et al., 1978), and their presence in the antibiotic-selected bacterial clones was confirmed by PCR using promoter- and gene-specific primers.
Plant transformation and selection of transgenics
An A. tumefaciens MP90 culture harbouring one of the binary vectors was grown for 1–2 d at 26–28 °C on a rotary shaker at 225 rpm to mid-log phase (OD600=1) in 15 ml liquid LB medium (Sambrook et al., 1989) supplemented with 100 mg l−1 kanamycin, 100 mg l−1 rifampicin, and 10 mg l−1 gentamicin. The bacterial cells were collected by centrifugation at 2000 g for 10 min, and resuspended to an OD600=0.4–0.5 in liquid tobacco regeneration medium (TRM) before plant inoculation. The TRM contained MS salts (Murashige and Skoog, 1962), MS or B5 vitamins (Gamborg et al., 1968), 20 g l−1 sucrose, 900 mg l−1 MES, 0.1 mg l−1 NAA, and 1 mg l−1 of either BA or ZR, at pH 5.7.
Fully developed young tobacco leaves were submerged in liquid TRM, and cut into 1 cm2 explants. After precultivation on liquid TRM for 2 d at 24 °C at low light intensity (5 μmol m−2 s−1), the explants were incubated with a fresh Agrobacterium solution (prepared as described above) for 1 h with slow shaking, then blotted with sterile filter paper to remove the excess bacteria, and placed with the adaxial side up on antibiotic-free TRM solidified with 0.4% (w/v) agarose. After 3–4 d of cultivation at 24 °C in low light, the infected explants were washed twice for 1 h with liquid TRM containing 1 g l−1 carbenicillin, and placed (three explants per plate) on to selective TRM containing 100–200 mg l−1 kanamycin, 500 mg l−1 carbenicillin, and 400 mg l−1 cefotaxime. The explants were cultivated at 24 °C with a 16 h light photoperiod (60 μmol m−2 s−1) and transferred to fresh antibiotics-containing TRM every 2 weeks. Regenerated shoots (1–1.5 cm high) were rooted in the hormone-free medium (see ‘Plant material’) supplemented with 100 mg l−1 kanamycin, 500 mg l−1 carbenicillin, and 400 mg l−1 cefotaxime.
PCR analysis of transgenic plants
Plant DNA was isolated from tobacco leaves using a GenElute™ Plant Genomic DNA Kit (Sigma, USA). PCR was carried out in 50 μl reaction mixtures containing 200 ng of plant DNA, Taq PCR Master Mix (Qiagen, USA) and specific primers, with manual hot start and the following parameters: 94 °C for 3 min, then 30 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for either 60 s (win3.12T-CEMA plants) or 90 s (win3.12T-GUS plants), followed by a final 10 min incubation at 72 °C. Primers for plants with the pwin3.12T-CEMA construct amplified the 90 bp full-length CEMA gene: forward primer 5′CEMA (5′-ATGAAATGGAAACTGTTCAAG-3′, 21-mer) and reverse primer 3′CEMA (5′-TTACTTAGTTAGCTTCAG-3′, 18-mer). In addition, forward primer 5′WIN (Yevtushenko et al., 2004) and reverse primer 3′CEMA were used to confirm the correct promoter–transgene fusion (928 bp amplified DNA fragment). Primers for plants with the pwin3.12T-GUS construct amplified the 1812 bp full-length GUS gene, and have been described previously (Yevtushenko et al., 2004).
Southern blot analysis
Four micrograms of tobacco DNA from each line were digested with 200 U of HindIII in a 200 μl reaction mix, concentrated, electrophoresed in a 1% (w/v) agarose gel, and transferred to a Biodyne B nylon membrane (Pall, USA). Prehybridization (3 h) and hybridization (overnight) were carried out at 65 °C in 10 ml of PerfectHyb™ Plus hybridization buffer (Sigma, USA) supplemented by 100 μg ml−1 of sheared, denatured salmon testis DNA (Sigma, USA). A 823 bp win3.12T promoter was labelled with [α-32P] dCTP to a specific activity of >109 cpm μg−1 by random priming, and used at 4×106 cpm of purified probe in 1 ml of hybridization solution. After hybridization, the membrane was washed twice with constant agitation in 2× SSC, 0.1% SDS at RT for 5 min, twice in 1× SSC, 0.1% SDS at 65 °C for 10 min, once in 0.5× SSC, 0.1% SDS at 65 °C for 10 min, and then exposed to X-ray film (Kodak BioMax) overnight at −80 °C using an intensifying screen. The membrane was also analysed with a PhosphorImager (Molecular Dynamics, USA).
Northern blot analysis
Total RNA was isolated from tobacco leaves using RNeasy® Plant Mini Kit (Qiagen, USA). The RNA samples (30 μg) were denatured and electrophoresed in a 1.2% formaldehyde agarose gel according to RNeasy® Mini Handbook protocol, checked for equal loading and RNA integrity by ethidium bromide staining, and transferred to a Biodyne B nylon membrane (Pall, USA) by capillary blotting. Prehybridization and hybridization were carried out at 65 °C using the PerfectHyb™ Plus hybridization buffer (Sigma, USA) as described in ‘Southern blot analysis’. To maximize the sensitivity of hybridization, the antisense DNA strand of the CEMA gene was 32P-labelled by linear PCR amplification using Strip-EZ™ PCR kit (Ambion, USA) and reverse primer 3′D/CEMA (5′-GGGGATCCTTACTTAGTTAGC-3′, 21-mer). The underlined part of the primer corresponds to the 3′ end CEMA region. An XbaI/EcoRI DNA fragment containing the full-length CEMA gene at the 5′ end was used as a template for run-off PCR probe synthesis. The PCR programme was as in ‘PCR analysis of transgenic plants’, except that the number of cycles was 40. The probe was purified from unincorporated nucleotides with a NICK™ column (Pharmacia Biotech, USA) and used at concentration of 2×106 cpm in 1 ml of the hybridization solution. After hybridization, the membranes were washed twice with constant agitation in 2× SSC, 0.1% SDS at RT for 5 min, twice in 1× SSC, 0.1% SDS at 65 °C for 10 min, and then exposed to X-ray film (Kodak BioMax) at −80 °C using an intensifying screen.
Antifungal activity of purified CEMA
The antifungal activity of CEMA peptide in vitro was quantified by determining the number of conidia germinated in the presence of the peptide. Fusarium solani was cultivated on a medium containing 10% (v/v) V8® juice, 5 mM MES, 15 g l−1 agar Difco (pH 6.4), at RT in low light. Conidia were collected aseptically by adding sterile dH2O to 1-week-old mycelium and gently scrubbing it with a spatula, then purified by filtration through sterile mesh, and adjusted to 105 conidia ml−1 in a 50-fold diluted potato dextrose broth (PDB; Difco, USA). The conidial suspension was dispensed into 100 μl aliquots in 96-well microplates, and incubated with different concentrations of CEMA peptide at RT in low light. The experiments were performed with three replications of each CEMA concentration. Germination of conidia was determined after 24 h of incubation, using a haemocytometer and an inverted light microscope. Conidia were considered germinated only if the newly grown hyphae were at least twice the length of the conidia (Jacobi et al., 2000).
CEMA phytotoxicity assay
Tobacco protoplasts were isolated from leaves of non-transgenic plants as described by Medgyesy et al. (1980) using a maceration solution of 0.2% (w/v) Cellulase (Sigma, USA), 0.1% (w/v) Driselase (Sigma, USA), 0.4 M sucrose, 0.1 M glycine, 10 mM CaCl2, and 10 mM MES (pH 5.6). The protoplasts were cultivated at density of 105 ml−1 in a thin layer of liquid 7p medium (Kao and Michayluk, 1975). After 3 d of cultivation at 24 °C in low light (5 μmol m−2 s−1), the cell culture was challenged with different concentrations of CEMA peptide. The effect of CEMA phytotoxicity was observed with an inverted light microscope: dividing cells and cells with spherical shape without any sign of cytoplasm degradation were defined as viable.
Transgenic plant wounding and tissue sampling
Mechanical wounding of tobacco leaves and tissue sampling for RNA and protein extractions were done as described earlier (Yevtushenko et al., 2004).
Plant resistance bioassays
For an in vitro regeneration bioassay, tobacco leaf explants were placed on antibiotic-free TRM (three 1 cm2 explants in each plate) and subjected to F. solani infection by placing two 1 cm2 agar blocks of freshly grown mycelia at an equal distance from the explants. The explants were co-cultivated with F. solani in a 16 h light photoperiod (60 μmol m−2 s−1) at 24 °C for several weeks. The evaluation of tissue resistance was based on its ability to remain green, form calli, and regenerate shoots in the presence of the pathogenic fungus.
For the bioassay of whole plants, two 1 cm2 agar blocks of the F. solani mycelia were placed 2 cm from the stem of a 4–5-week-old tobacco plant grown aseptically on antibiotic-free medium (see ‘Plant material’), and cultivated for several weeks. The plants were observed daily for disease symptoms, and scored for resistance using the following criteria: susceptible, if the plants developed severe wilt symptoms such as brown stems and dehydrated leaves within the same time period as non-transgenic plants; moderately resistant, 2–3 times longer period to develop the wilt symptoms; resistant, 4–5 times longer; and highly resistant, more than 6 times longer period before any disease symptoms appeared. The bioassays were replicated three to five times with each transgenic line.
To study pathogen-induced CEMA expression, two 1 cm2 agar blocks of the fungal mycelia were placed 2 cm from the stem of a young well-developed tobacco plant grown in a Magenta GA7 vessel (Magenta, USA), and cultivated for several days. One to two days after fungal mycelia reached the plant stem, all leaves were collected for RNA extractions.
Antifungal activity of leaf extracts
Transgenic and control tobacco plants were co-cultivated with F. solani as described above. One to two days after the fungus contacted the stem, all the leaves were collected and ground to a fine powder in liquid nitrogen with no buffer added. Ground tissue was centrifuged at 14 000 g for 10 min at 4 °C to pellet the cellular debris, and the supernatant containing total leaf extract was mixed with a protease inhibitor cocktail (Sigma, USA). F. solani conidial suspension was adjusted to a density of 106 conidia ml−1 in a 5-fold diluted PDB (Difco, USA), mixed 1:9 with the leaf extract, and incubated at RT in low light. The percentage of germinated conidia was assessed after 24 h of incubation, as described in ‘Antifungal activity of purified CEMA’. The assay was replicated at least three times.
GUS assay
Quantitative fluorometric assays of GUS activity in leaf samples, harvested both before and after mechanical wounding, were performed as described earlier (Jefferson et al., 1987; Gallagher, 1992) by incubation of the extracts (20 μg protein) with 2 mM 4-methyl-umbelliferyl-β-D-glucuronide (MUG) in a lysis buffer for 60 min at 37 °C. GUS activity was calculated as pmol of 4-methyl-umbelliferone (4-MU) produced min−1 mg−1 of soluble protein.
Results
Antifungal activity of CEMA in vitro
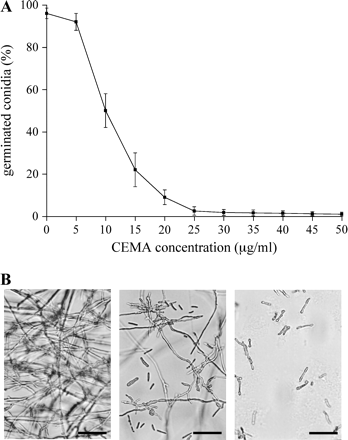
Since no data on antifungal activity of CEMA were available, this synthetic peptide was tested in vitro against the plant-pathogenic fungus F. solani. Salts and PDB above diluent threshold concentrations inhibit the effect of antimicrobial peptides on the germination of conidia (Jacobi et al., 2000), therefore all these quantitative assays were performed in a 50-fold diluted modified PDB. A dose–response curve was obtained after 24 h incubation of fungal conidia with CEMA (Fig. 1A), showing that as little as 10 μg ml−1 peptide was sufficient to inhibit germination of 50% conidia (IC50). The minimal inhibitory concentration (MIC) of CEMA, defined here as the lowest peptide concentration that reduced the germination of conidia by at least 95%, was 23 μg ml−1. No fungal growth was usually detected above this concentration. Even if some conidia (<5%) germinated above the MIC, they had such severe hyphal abnormalities that their growth was terminated shortly after germination. Significant changes in the hyphal morphology were also observed at concentrations that only partially inhibited the germination of conidia (<IC50). It included abnormal hyperbranching of germinated conidia and forming condensed, short hyphae of only a few cells in length, compared with thin, well-extended mycelial growth in controls (Fig. 1B). Thus, even sublethal concentrations of CEMA inhibited fungal growth.
Antifungal activity of CEMA peptide in vitro. (A) Effect of CEMA on germination of F. solani conidia after 24 h of incubation. Values are the means of three experiments; bars are standard errors. (B) Hyphal growth anomalies in the presence of CEMA. Photographs were taken 24 h after incubation of F. solani conidia without CEMA (control, left photo), with 10 μg ml−1 CEMA (central photo), and with 30 μg ml−1 CEMA (right photo). Bar: 100 μm.
Interestingly, 2 d after the assessment of CEMA activity in vitro, more germination of conidia and mycelial growth was observed at concentrations previously determined as inhibitory for this fungus. Similar observations were reported in experiments with other AMPs (Qui et al., 1995; Cavallarin et al., 1998). The antifungal potency of CEMA did not decrease over time only at concentrations equal to or above the MIC. Addition of proteinase inhibitors alone did not inhibit fungal growth, but their presence in the medium with CEMA prevented the observed loss of peptide activity, indicating that degradation of CEMA was caused by secreted fungal proteinases.
No antifungal activity was detected after incubation of CEMA with proteinase K, and the rate of germination of conidia (>95%) was similar to that in control experiments without CEMA, or with proteinase K only. Similar to other AMPs (Jacobi et al., 2000), germination of conidia exposed to CEMA without proteinase K treatment was strongly inhibited, confirming that the CEMA peptide itself, rather than any other non-protein compounds, was responsible for the observed antifungal activity.
Evaluation of CEMA phytotoxicity
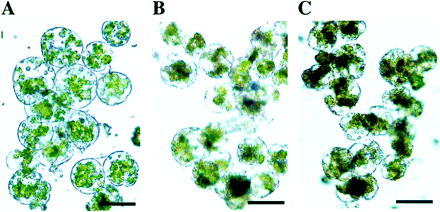
To be useful for engineering disease-resistant plants, CEMA should have no toxic effects on host plants. Because AMPs interact with the cell membrane, a bioassay of plant protoplasts cultivated in liquid medium presents a highly sensitive indicator of peptide phytotoxicity. To test the effect of CEMA on plants, tobacco mesophyll protoplasts were cultivated with different amounts of peptide, ranging from 0 to 100 μg ml−1 (Fig. 2). The threshold for CEMA concentrations toxic to tobacco cells was found to be 30–35 μg ml−1. No visible effects on protoplast viability and cell divisions were observed at peptide concentrations below 30 μg ml−1. At 30 μg ml−1, CEMA started to inhibit cell divisions and some cells (less than 50%) turned brown and died within 24 h. Visually, the toxic effect of CEMA on plant cells included a change of cell shape and colour (browning of chloroplasts), with the cytoplasm becoming dark and separated from the cell wall indicating cell death. When 40 μg ml−1 CEMA was added to protoplast culture, no cells survived longer than 24 h. Overall, the concentration of peptide that affected plant cell viability was about 3-fold higher than that required to inhibit germination of conidia by 50%, and 7-fold higher than IC50 determined for bacterial pathogens (Osusky et al., 2000).
Effect of CEMA peptide on the viability of plant protoplasts in vitro. Tobacco mesophyll protoplasts were cultivated for 5 d before addition of the peptide. Photographs were taken 24 h after incubation of plant protoplasts without CEMA (A), with 30 μg ml−1 CEMA (B), and 50 μg ml−1 CEMA (C). Bar: 50 μm.
Production of transgenic plants
Inhibition of fungal growth by CEMA in vitro, combined with its low phytotoxicity, encouraged the evaluatation of the efficacy of this AMP for providing pathogen control in planta. A plant transformation vector with transcriptional fusion between the gene encoding CEMA and an 823 bp wound-inducible promoter win3.12T from poplar was constructed (Fig. 3B) and introduced into N. tabacum L. cv. Xanthi via Agrobacterium-mediated transformation. As a control for promoter activity, tobacco was transformed with a construct designed to express β-glucuronidase (GUS) reporter gene from the win3.12T promoter. Stable transgene integration into plants regenerated on selective medium was confirmed with PCR and Southern analyses (Fig. 3C, D), indicating that one to four copies of the transgene were maintained in the plant genome (Fig. 3D). All transgenic plants had the normal phenotype of tobacco, with no indication of cytotoxicity due to expression of the CEMA gene. Eight lines with win3.12T-CEMA construct and six lines with win3.12T-GUS construct were randomly selected among the transgenic plants for further analyses.
(A) Nucleotide and amino acid sequences of the cecropin A-melittin (CEMA) hybrid. The cecropin A part of the CEMA is shown in light, and the melittin part is in bold. (B) A schematic presentation of the pwin3.12T-CEMA construct for tobacco transformation. The relative positions of the win3.12T promoter, the CEMA gene, and restriction sites for cloning are shown. The positions of PCR primers for DNA analyses are indicated by arrows. PolyA (NOS-t) is a poly-adenylation sequence of the nopaline synthase gene. (C) PCR analysis of DNA isolated from eight transgenic tobacco lines. Fragments of 928 bp were generated using promoter- and CEMA-specific primers (5′-WIN and 3′-CEMA, respectively), and they indicated both the presence of the transgene (90 bp) and the correct promoter–transgene fusion. Lanes 1 and 13: 1 kb DNA ladder. Lane 2: PCR mix without template DNA. Lane 3: plasmid pwin3.12T-CEMA. Lane 4: non-transgenic tobacco. Lanes 5–12: transgenic tobacco lines Twc1, Twc2, Twc3, Twc6, Twc7, Twc10, Twc11, and Twc12, respectively. (D) Southern blot analysis of transgenic plants. Tobacco DNA was digested with HindIII, electrophoresed and hybridized with a 32P-labelled win3.12T promoter probe. The number of bands reflects the number of transgene insertions. Transgenic line Twc6 with a band of higher signal intensity contains four transgene copies, as determined by a Molecular Dynamics densitometer. Lane 1: 1 kb DNA ladder. Lane 2: non-transgenic tobacco. Lanes 3–10: transgenic tobacco lines Twc1, Twc2, Twc3, Twc6, Twc7, Twc10, Twc11, and Twc12, respectively. Molecular weight DNA markers are shown on the left.
Wound-inducible activity of the win3.12T promoter
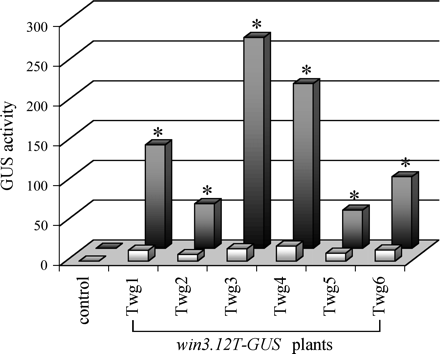
Because a truncated version, containing only 823 bp of the original wound-inducible promoter (Hollick and Gordon, 1993, 1995), was used in this study, there was concern initially as to whether the win3.12T length was sufficient to maintain its wound-regulated activity. Therefore, a GUS reporter gene fusion was used to evaluate activity of the win3.12T in response to wounding. Twenty hours after mechanical wounding of leaves, all transgenic lines showed 4–17-fold increase in GUS accumulation in the uninjured part of the wounded leaf, compared with that in the same leaves before treatment (Fig. 4). A significant response of the win3.12T promoter to wounding was also confirmed by quantitative RT-PCR of DNase-treated leaf RNA from a selected transgenic plant, showing an almost 10-fold increase in accumulation of GUS mRNA after wounding. By contrast, the activity of the win3.12T promoter in unwounded leaves was very low, with GUS expression only slightly higher than endogeneous GUS-like fluorescence in non-transgenic plants (background). These results were similar to wound-regulated activity of the win3.12T promoter in transgenic potato (Yevtushenko et al., 2004), indicating that the truncated poplar promoter was sufficient to confer a local response to wound stimuli in both heterologous hosts (tobacco and potato).
Wound-induced activity of win3.12T promoter in transgenic tobacco. Bar graph represents GUS activities in leaves of transgenic plants containing the win3.12T-GUS construct, both before (light bars) and 20 h after (dark bars) mechanical wounding. Mean values of GUS activities ±SE in pmol MU mg−1 protein min−1 were estimated in three to five plants of each transgenic line. Control denotes non-transgenic plants. Asterisks indicate win3.12T-driven GUS activities in wounded leaves that are significantly different (P <0.05, by Student's t-test) from the GUS activities in the same leaves before wounding.
Win3.12T-driven CEMA accumulation and transgene copy number
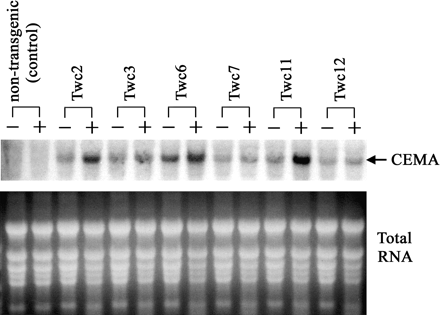
To examine whether the introduced CEMA gene is expressed in plants, the accumulation of CEMA transcripts was determined by northern blot analysis of leaf RNA isolated both before and after wounding. The result showed CEMA expression in all transgenic lines. It confirmed a higher level of transgene expression after wounding and also showed large variation in wound-induced transcript accumulation among the lines (Fig. 5). Comparison of these data with Southern analysis (Fig. 3D) revealed that wound-induced accumulation of CEMA mRNA was proportional to the number of transgene insertions: the highest CEMA expression was detected in lines Twc2, Twc6, and Twc11 that contained 3–4 transgene copies, whereas the lines with lowest transgene expression (Twc7 and Twc12) had only 1 transgene insertion.
Northern blot analysis of CEMA mRNA accumulation in tobacco. Total RNA was prepared from leaves of transgenic plants, both before (−) and 18 h after (+) mechanical wounding. RNA samples (30 μg each) were separated by denaturing formaldehyde-agarose gel electrophoresis, blotted, and hybridized with a 32P-labelled CEMA probe. Ethidium bromide stained ribosomal RNA bands (lower picture) are shown as loading controls.
In vitro regeneration bioassay for pathogen-resistant tissues
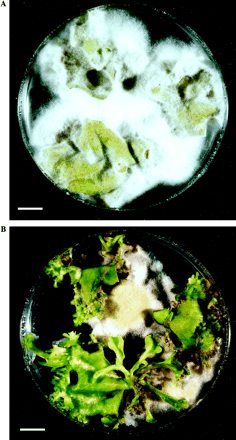
To screen transgenic lines for resistance to pathogens, a simple and efficient procedure was developed based on the ability of resistant tissue to form calli and regenerate plants in the presence of a pathogen. For this purpose, leaf explants were cultivated on antibiotic-free tobacco regeneration medium containing the pathogenic fungus F. solani, and their regeneration response was observed. This Fusarium sp. grew quickly, and in 3–4 d mycelia had spread completely over the medium in the Petri dish in control experiments. Using this bioassay, a dramatic difference was found in morphogenetic capacity among the transgenic lines. After 2 weeks of co-cultivation with the fungus, the calli formation and plant regeneration were most pronounced in lines Twc6 and Twc11 that remained healthy and green, producing multiple shoots on each explant (Fig. 6B). Moreover, the fungal growth ceased in Petri dishes containing transgenic tissues, and mycelia changed colour from normal white to yellow-brown and even formed a clear zone next to the proliferating plant tissue. It is likely that the transgenic plant tissue released the antimicrobial peptide (CEMA) into the cultivation medium, thereby inhibiting fungal growth. By contrast, the leaf explants from non-transgenic tobacco (control) were completely invaded by fungus with no signs of calli formation, and died within the first week of cultivation (Fig. 6A). Some transgenic lines (Twc7 and Twc12) were found to be almost as susceptible as the control, whereas other lines (Twc1, Twc2, Twc3, and Twc10) showed moderate morphogenetic response, varying from calli growth with no shoots to a few regenerated plantlets per experiment. Interestingly, the cultivation of the explants with F. solani for a longer period (over 3 weeks) resulted in the gradual inhibition and death even of those transgenic lines that were previously determined as resistant; however, it was not due to lack of sufficient resistance but rather a result of nutrient depletion and, perhaps, medium intoxication by metabolites from the dying fungus. Overall, the observed spectrum of morphogenetic capacity among the lines strongly correlated with data from northern and Southern analyses: the best regeneration response was detected in the lines that accumulated the highest amount of wound-induced CEMA mRNA (Fig. 5) and had higher gene copy number (Fig. 3D).
In vitro plant regeneration bioassay. Leaf explants of non-transgenic (A) and CEMA-expressing transgenic line Twc6 (B) are shown on antibiotic-free tobacco regeneration medium after 16 d of co-cultivation with F. solani. Bar: 1 cm.
Plant resistance to F. solani and pathogen-induced CEMA expression
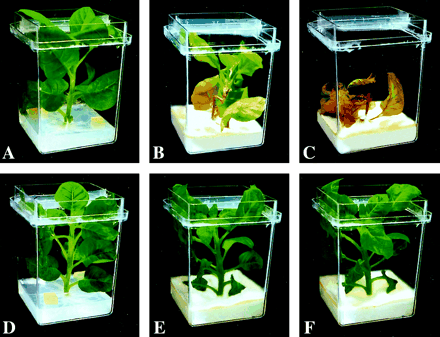
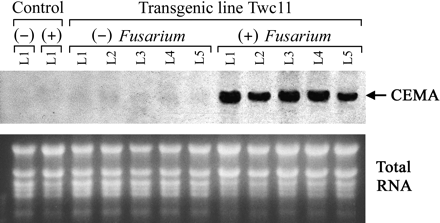
To evaluate whole plants for resistance to fungal infection, stringent biotests were conducted by growing the plants in vitro in the presence of F. solani. Because the plant medium contained a relatively high level of sucrose (20 g l−1), it promoted a high rate of continuous mycelial growth, thereby creating an extremely harsh environment for plant survival. Plant assessment was based on the extent of stem discolouration, wilt symptoms, and days of survival after fungal infection. One to two days after mycelium contacted the plant stem, all non-transgenic plants displayed browning and necrosis of the entire vascular system, resulting in a severe wilt and death within less than 1 week (Fig. 7A, B, C). By contrast, transgenic lines Twc6 and Twc11 survived longer than 2 weeks indicating significantly enhanced resistance to Fusarium infection (Fig. 7D, E, F). Among the other lines, Twc7 and Twc12 were susceptible and survived no longer than non-transgenic controls, Twc1, Twc3, and Twc10 were moderately resistant showing disease symptoms 3–4 d later compared with the controls, and Twc2 was resistant, surviving longer than 7 d. The observed differences in disease resistance corresponded with the in vitro regeneration bioassays and reflected the expression level of CEMA transcripts in response to wounding. Northern blot analysis of a representative line showed high level of CEMA mRNA accumulation in leaves after fungal infection (Fig. 8). Moreover, the pathogen-induced expression of CEMA was detected in all leaves of the plant, confirming that the resistance against F. solani was associated with the strong systemic activity of the win3.12T promoter in response to fungal infection.
Plant resistance to the pathogenic fungus F. solani. Two 1 cm2 agar blocks of the fungal mycelia were placed 2 cm from the stem of a well-developed tobacco plant grown in vitro, and cultivated for several weeks. The upper row represents non-transgenic (control) plants; the lower row represents CEMA-expressing transgenic line Twc11. (A, D) Start of co-cultivation with F. solani. (B, E) 6 d later. (C, F) 11 d later.
Pathogen-induced, systemic accumulation of CEMA transcripts. Total RNA was prepared from leaves of transgenic line Twc11 in the absence of treatment (−), and after the plant was subjected to Fusarium infection (+). L1, L2, L3, L4, and L5 denote different leaves of the plant, counted upward from the stem base, with L1 being the lowest leaf. Control indicates non-transgenic plant. RNA samples (20 μg each) were separated by denaturing formaldehyde–agarose gel electrophoresis, blotted, and hybridized with a 32P-labelled CEMA probe. Ethidium bromide-stained ribosomal RNA bands (lower picture) are shown as loading controls.
Antifungal activity of leaf extracts from transgenic plants
To confirm that the observed resistance to F. solani was due to biological activity of the CEMA peptide in plants and not because of natural defensive compounds produced by the plants, the antifungal activity of extracts from pathogen-infected tobacco plants was assessed. The extracts were treated with proteinase inhibitors. Among all transgenic plants, leaf extracts from the lines with highest CEMA expression (lines Twc6 and Twc11) had the strongest inhibitory effects on germination of F. solani, resulting in more than 70% reduction in the number of fungal colonies. Moreover, many germinated conidia developed hyphal growth anomalies similar to those observed in the in vitro bioassays with pure CEMA. Antifungal activities of leaf extracts from other lines varied, yet they were high enough (except lines Twc7 and Twc12) to be significantly different (P <0.05, by Student's t-test) from the corresponding non-transgenic controls. The number of germinated conidia in leaf extracts from non-transgenic plants was lower than that in water or PDB, indicating that probably some compounds produced by plants also contributed to the inhibition of fungal growth. Overall, these results were in agreement with the plant resistance bioassays and reflected the expression level of CEMA mRNAs among the lines.
Discussion
Expression of defensive genes from a promoter that is specifically activated in response to pathogen invasion or pest attack has distinct advantages for crop improvement. The application of a strong pathogen-inducible promoter to direct expression of a novel AMP, which resulted in successful engineering of plant resistance against fungal infection, has been demonstrated here for the first time. Previously it was shown that an 823 bp wound-inducible win3.12T promoter from hybrid poplar (Populus trichocarpa×P. deltoides) effected a strong systemic activity in aerial parts of potato in response to fungal infection (Yevtushenko et al., 2004). A strong pathogen-induced systemic activity of the win3.12T promoter observed here in another heterologous host (tobacco) indicates that the promoter sequence contains highly conserved regulatory elements activated by the same signalling mechanism in both plant species. To date, pathogen-inducible plant promoters are known to share the W-box motifs with the core consensus TGAC (for a review, see Eulgem et al., 2000). These W-boxes act as important cis elements that interact with elicitor-induced DNA-binding proteins of the WRKY family of transcription factors, and are required for promoter activation in response to elicitor treatment or pathogen infection. Five clustered copies of the W-box motif in the poplar win3.12T promoter were identified here. Experiments with the different promoter deletions are currently in progress to study whether these elements are essential for pathogen responsiveness in this system. Because the win3.12T promoter is also induced by mechanical wounding, it is still unclear whether this promoter contains specific wound-responsive regulatory elements, or its wound-induced activity is a result of crosstalk between wound and pathogen/elicitor signalling pathways.
In this work, the win3.12T-regulated CEMA expression in leaves was directly proportional to the number of transgene insertions. Similar correlation is observed in plants where the win3.12T promoter directed the expression of the GUS reporter gene (Yevtushenko et al., 2004). Other reports have shown that transgene copy number can have positive (Gendloff et al., 1990; Elomaa et al., 1995), negative (Finnegan and McElroy, 1994), or no effect (Peach and Velten, 1991) on transgene expression. Although an increased copy number has often been associated with transgene silence (for review see Finnegan and McElroy, 1994), the highest number of transgene insertions in plants in this study had no inhibitory effect on the accumulation of CEMA transcripts. Furthermore, because CEMA activity was decreased by the presence of proteinases in the in vitro bioassays, multiple copies of this gene might be required to compensate for protein degradation and to fight disease effectively. Therefore, the observed correlation between the number of insertions and transcript accumulation presents an advantage to predict and manipulate the win3.12T-driven transgene expression.
Previous studies on the expression of native AMPs, particularly cecropin B, have shown contradictory results on disease resistance in plants due to the degradation of the peptide by host proteases (Jaynes et al., 1993; Hightower et al., 1994; Florack et al., 1995; Huang et al., 1997). Fortunately, due to their short peptide lengths, most native AMPs can easily be modified based on the rules of peptide modelling. These new synthetic AMPs are much more powerful than their native counterparts, less sensitive to proteases, and have a low toxic effect on the host plant (Andreu et al., 1992; Powell et al., 1995; Dykes et al., 1998; Tiozzo et al., 1998; Jacobi et al., 2000). The novel synthetic chimeric peptide CEMA was tested here for the ability to provide disease resistance in tobacco. This peptide contains the appropriate sequence juxtaposition of N-terminal sections of cecropin A and melittin, and was designed for improved potency of the native cecropin A without the haemolytic effects of melittin (Hancock et al., 1992). It was shown that CEMA had a strong antifungal activity in vitro, inhibiting the germination of conidia at concentrations that were non-toxic to tobacco protoplasts. Intact plant tissues were even less susceptible to the inhibitory effect of CEMA than protoplasts (personal observations). Thus, in vitro data suggested that CEMA was an appropriate candidate to be expressed in plants for antifungal resistance, with a low probability of deleterious effects in plant cells. Similar antifungal activity in vitro against several agronomically important pathogens has been reported for other cecropin A-derived and cecropin A-melittin hybrid peptides (Cavallarin et al., 1998; Ali et al., 2000); however, no data on the activity of these AMPs in plants are available.
To evaluate the efficacy of CEMA in vivo, the CEMA-encoding gene was transcriptionally fused to the win3.12T promoter, and tested in transgenic tobacco plants. A high accumulation of CEMA transcripts was found in leaves of transgenic plants after mechanical wounding (mimicking pathogen invasion) or fungal infection, whereas in unstressed leaves it was hardly detectable. Moreover, the pathogen-induced CEMA expression was systemic, confirming the advantage of the win3.12T promoter over other promoters to elicit expression of a defensive transgene only when the plant is attacked by a pathogen. Assessment of plant survival in the presence of F. solani showed that host resistance depended on the amount of CEMA expressed: transgenic lines expressing the highest levels of CEMA were able to withstand fungal infection for more than 2 weeks, whereas the lines with low CEMA expression survived no longer than non-transgenic controls. The antifungal resistance of plants with the highest CEMA expression was strong and reproducible. Strong inhibition of the germination of conidia by leaf extracts from pathogen-infected transgenics confirmed that the CEMA peptide itself rather than other compounds was responsible for the enhanced resistance of the tobacco plants. All transgenic plants were healthy and showed no morphological or developmental abnormalities even in the lines with the highest CEMA mRNA accumulation. Thus, the expression level of CEMA in tobacco plants was sufficient to maintain its antifungal activity without any toxic effects to the transgenic host. Interestingly, resistance or susceptibility of plants towards Fusarium is at least partly determined by interactions occurring within xylem vessels (Rep et al., 2002). Although the sequence of CEMA used in this work was not preceded by a specific signal peptide designed to translocate the mature CEMA into intercellular spaces, the expression level of CEMA in selected plants was sufficient to elicit its antifungal action.
In the search for a quick, space-saving, and reliable screening of transgenic lines for pathogen-resistance, an efficient bioassay was developed based on the ability of resistant tissue to regenerate in the presence of a pathogen. Plant regeneration is extremely sensitive to any unfavourable factors in the surrounding environment: change in medium composition, pH, temperature, or the presence of pathogenic micro-organisms will immediately affect cell proliferation. Therefore it was hypothesized that if the transgenic lines generated a sufficient level of the antimicrobial peptide, they would have an advantage in morphogenetic capacity over susceptible lines when subjected to fungal infection. Indeed, only the lines with the highest CEMA expression remained green, formed calli and regenerated shoots when challenged with F. solani. Moreover, the morphogenetic capacity of the leaf explants reflected the large variation in CEMA expression among the lines. This regeneration bioassay can be adapted for screening of transgenics expressing other AMPs. More importantly, it permits the identification of transformed tissue without co-expression of a selectable marker gene, thus providing an opportunity to produce transgenic plants free of antibiotic-resistance markers. The latter is a great advantage for regulatory and public acceptance of transgenic crops.
Thus, the data obtained in this study are highly encouraging and suggest that pathogen-inducible expression of the cecropin A-melittin peptide gene, when controlled by the win3.12T promoter, provides an effective approach in engineering plant resistance against fungal disease. Further efforts are in progress to express other defence genes under regulation of the win3.12T promoter to test the generality of this study for crop improvement.
Present address: The Huntsman Marine Science Centre, 1 Lower Campus Road, St Andrews, New Brunswick, E5B 2L7 Canada.
Abbreviations: CEMA, cecropin A-melittin hybrid peptide; AMP, antimicrobial peptide; GUS, β-glucuronidase; TRM, tobacco regeneration medium; PDB, potato dextrose broth; MIC, minimal inhibitory concentration.
This research was supported by grants from the National Center of Excellence (NCE), Canadian Bacterial Diseases Network (CBDN), and Advanced Food and Materials Network (AFMN) to S.M.
References
Ali GS, Reddy ASN.
Allefs SJHM, Florack DEA, Hoogendoorn C, Stiekema WJ.
Andreu D, Ubach J, Boman A, Wåhlin B, Wade D, Merrifield RB, Boman HG.
Cavallarin L, Andreu D, San Segundo S.
Chakrabarti A, Ganapathi TR, Mukherjee PK, Bapat VA.
DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H.
De Lucca AJ, Bland JM, Grimm C, Jacks TJ, Cary JW, Jaynes JM, Cleveland TE, Walsh TJ.
Dykes GA, Aimoto S, Hastings JW.
Elomaa P, Helariutta Y, Griesbach RJ, Kotilainen M, Seppänen P, Teeri T.
Eulgem T, Rushton PJ, Robatzek S, Somssich IE.
Florack D, Allefs S, Bollen R, Bosch D, Visser B, Stiekema W.
Gallagher SR.
Gamborg OL, Miller RA, Ojima K.
Gao A, Hakimi SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CMT.
Gendloff EH, Bowen B, Buchholz WG.
Hancock REW, Brown MH, Piers K.
Hancock REW, Lehrer R.
Hightower R, Baden C, Penzes E, Dunsmuir P.
Hollick JB, Gordon MP.
Hollick JB, Gordon MP.
Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J.
Huang Y, Nordeen RO, Di M, Owens LD, McBeath JH.
Jacobi V, Plourde A, Charest PJ, Hamelin RC.
Jaynes JM, Nagpala P, Destefano-Beltran L, Huang JH, Kim JH, Denney T, Cetiner S.
Jefferson RA, Kavanagh TA, Bevan MW.
Kao KN, Michayluk MR.
Medgyesy P, Menczel L, Maliga P.
Mentag R, Luckevich M, Morency M-J, Séguin A.
Murashige T, Skoog F.
Nicolas P, Mor A.
Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S.
Powell WA, Catranis CM, Maynard CA.
Peach C, Velten J.
Qui X, Wu Y, Jaynes J, Goodwin P, Erickson LR.
Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, Speijer D, Back JW, de Koster CG, Cornelissen BJC.
Sambrook J, Fritsch EF, Maniatis T.
Sharma A, Sharma R, Imamura M, Yamakawa M, Machii H.
Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG.
Tiozzo E, Rocco G, Tossi A, Romeo D.
Yevtushenko DP, Sidorov VA, Romero R, Kay WW, Misra S.




Comments