-
PDF
- Split View
-
Views
-
Cite
Cite
M. Hallin, O. Denis, A. Deplano, R. De Mendonça, R. De Ryck, S. Rottiers, M. J. Struelens, Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey, Journal of Antimicrobial Chemotherapy, Volume 59, Issue 3, March 2007, Pages 465–472, https://doi.org/10.1093/jac/dkl535
Close - Share Icon Share
Abstract
Surveillance of hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA) infections has shown the emergence and spread of several epidemic MRSA clones over the past 10 years in Belgium. Whether these clones have been imported from abroad or else have arisen locally via staphylococcal cassette chromosome mec (SCCmec) acquisition by successful methicillin-susceptible S. aureus (MSSA) clones is unknown.
We determined by PFGE, spa typing, multi-locus sequence typing (MLST) and agr group analysis the genetic relatedness of 103 MSSA and 511 MRSA strains from a national survey of patients admitted to 112 Belgian hospitals in 2003.
The 103 MSSA strains presented very diverse genetic backgrounds, they were distributed into 40 distinct PFGE types and clustered in 15 distinct MLST CCs. Up to 45% harboured the same genotype as five major epidemic HA-MRSA clones. These MRSA clones all harbour a type IV SCCmec element.
These findings are consistent with multiple recent acquisitions of the more mobile type IV SCCmec by MSSA and suggest that certain genetic backgrounds are conferring a selective advantage, regardless of the resistance profile. However, since the predominant MSSA and MRSA lineages identified in Belgium are disseminated worldwide, importation of epidemic MRSA strains remains an alternative hypothesis.
Introduction
Staphylococcus aureus is a leading cause of human disease in the hospital setting as well as in the community accounting for a wide range of diseases from superficial skin infections to life-threatening conditions such as bacteraemia, endocarditis, pneumonia or toxic shock syndrome.1 Resistance to methicillin (and related β-lactam drugs) first appeared in 1961, shortly after its introduction into clinical practice.2 Since then, methicillin-resistant S. aureus (MRSA) has become a major cause of nosocomial infections in many countries all over the world.
The mecA gene, conferring methicillin resistance, is carried by a mobile exogenous genetic element called staphylococcal cassette chromosome mec (SCCmec), probably first acquired by horizontal transfer from coagulase-negative staphylococcal species.3,4 Six major types of SCCmec differing in size and content have been described and characterized according to the type of recombinase genes (ccr) and the class of mec complex they carry.5,6 Multi-locus sequence typing (MLST) of seven housekeeping genes, combined with SCCmec typing, applied to a large international S. aureus strains collection led to the conclusion that MRSA arose on multiple occasions (at least 20 times) from successful methicillin-susceptible S. aureus (MSSA) clones, leading to a relatively small number of pandemic MRSA clones worldwide.7
In Belgium, the proportion of methicillin-resistant strains among clinical S. aureus isolates has risen from 21% in 19948 to 29% in 2003.9 Molecular epidemiology surveys have shown that this recent increase occurred simultaneously with the emergence followed by wide diffusion of two novel epidemic MRSA clones (PFGE B2-ST45-SCCmec type IV and PFGE A20-ST8-SCCmec type IV). These clones, as well as three other recent but less diffused clones (A21-ST8-SCCmec type IV, C3-ST5-SCCmec type IV, L1-ST22-SCCmec type IV) are susceptible to several non-β-lactam antibiotics and carry the SCCmec type IV.10 Like in other European countries,11–13 these MRSA clones have progressively supplanted the multidrug-resistant PFGE A1-ST247-SCCmec I ‘Iberian’ clone which had been predominant in Belgian hospitals during the 1980s and early 1990s.7,10,14
Belgium, like other countries, is also facing the emergence of MRSA infections acquired in the community (CA-MRSA). Interestingly, CA-MRSA strains found in Belgium have a distinct genetic background from hospital-acquired (HA)-MRSA and share the same two characteristics with new epidemic HA-MRSA clones: limited resistance to non-β-lactam antibiotics and SCCmec type IV.15
Those major changes in the epidemiology of MRSA raise the question of the origin of emergent clones harbouring SCCmec IV. Indeed, type IV is the smallest SCCmec type and the most widely distributed SCCmec type among S. aureus lineages. This ‘ecological success’ has been linked to a lower ‘fitness cost’.7,16 To test the hypothesis of new HA- and CA-MRSA clones arising locally via SCCmec IV acquisition by successful MSSA clones, the authors explored the genetic background of MSSA strains in patients admitted to Belgian acute-care hospitals in 2003 and compared them with MRSA clones currently prevalent in this setting.
Materials and methods
Bacterial strains
From January to December 2003, the national Reference Laboratory asked microbiological laboratories serving all Belgian acute-care hospitals (n = 180) to collect one MSSA and five non-duplicate MRSA strains per hospital site, recovered in hospitalized patients from any body site. The strains were sent to the Reference Laboratory with a case report form describing the following demographic data: patient age and sex, type of specimen, and category of hospital unit. The characterization by PFGE, MLST and SCCmec typing of MRSA strains collected during this survey (n = 511) as well as their genetic distribution was described previously.17
DNA extraction
DNA was extracted as described by Ünal et al.18 Briefly, each isolate was cultured for 24 h on Columbia agar with 5% sheep blood, then successively incubated with lysostaphin and proteinase K, boiled and finally harvested. This lysate was used as DNA template in all PCRs described below.
Identification and characterization of oxacillin resistance
All isolates were confirmed by PCR for 16S rRNA, nuc and mecA genes by PCR as previously described.19
Molecular typing methods
SmaI restriction of genomic DNA was separated by PFGE in a CHEF-Mapper system (Bio-Rad Laboratories, Nazareth, Belgium) as previously described.14 The similarity of macrorestriction patterns was determined both by visual comparison and by computer matching using BioNumerics software version 4.0 (Applied Maths, Ghent, Belgium). Restriction fragments in the size range between 36 and 700 kb were analysed by Dice similarity coefficient with a position tolerance set to 0.8%. A dendrogram of similarity was built using the unweighted pair-group method using arithmetic averages (UPGMA). Patterns differing by fewer than seven fragments were considered to belong to the same group (represented by a capital letter), and those differing by 0–3 fragments to the same type (represented by a number).14 PFGE types present in more than two hospitals were classified as ‘epidemic types’.
spa typing was performed as described by Harmsen et al.20 on one randomly selected strain of every epidemic PFGE type. spa types were determined with the Ridom StaphType software version 1.3 (Ridom GmbH, Würzburg, Germany) and analysed by the BURP algorithm with the default parameters: spa types shorter than 5 repeats are considered as non-typeable and spa types belong to the same group if cost is less or equal 6 (EDSI model21).
agr type was determined by multiplex PCR as described by Gilot et al.22 on the same sub-sample as spa typing.
Multi-locus sequence typing (MLST) was performed as described by Enright et al.23 on a representative set of strains belonging to major MSSA PFGE groups (n = 20). The sequence types were determined with the MLST database accessible via http://www.mlst.net. Sequences were aligned and maximum parsimony trees were built with BioNumerics 4.0 (Applied Maths, Ghent, Belgium).
Toxin gene detection
The presence of Panton-Valentine leucocidin (PVL) (lukS-PV and lukF-PV) and TSST-1 genes was tested by duplex PCR as described by Lina et al.24 on all MSSA strains and on a random sub-sample of isolates belonging to the nine most frequent MRSA PFGE types (n = 156).17
Results
Hospital participation and bacterial strains
One hundred and twelve hospitals participated (62% of all acute care hospitals). They were located in Brussels (n = 16), Flanders (n = 57) and Wallonia (n = 39). A total of 547 MRSA and 112 MSSA isolates were collected of which 511 strains were confirmed by PCR as MRSA (93%) and 103 as MSSA (92%). The remaining 35 isolates were excluded from the study as they were either misidentified staphylococci (n = 20) or did not grow on subculture (n = 15).
Demographic data
Six hundred and five patient report forms were available (510 from MRSA strains, 95 from MSSA strains). Patients with MRSA infection or colonization were older than patients with MSSA infection (P < 0.01, Table 1). They had a similar sex ratio. Patients' distribution in various hospital care units were similar in both groups (P = 0.06, Table 1). MRSA and MSSA strains were recovered from diverse specimen types, but MRSA strains were significantly more often recovered from screening cultures than MSSA (P < 0.01) and a larger proportion of MSSA strains were recovered from blood cultures (P < 0.01, Table 1).
Sex, age, type of specimen and category of hospitalization unit of 510 patients with MRSA and 95 patients with MSSA infection or colonization
| . | Patients with MRSA . | Patients with MSSA . | P value . |
|---|---|---|---|
| Sex ratio (M/F) | 0.8 | 1.1 | 0.15* |
| Median age, years (range) | 78 (0–97) | 67 (1–96) | < 0.01** |
| Hospitalization units | 0.06* | ||
| medical wards | 32% | 28% | |
| intensive care units | 10% | 13% | |
| surgical wards | 18% | 22% | |
| geriatric wards | 26% | 15% | |
| other | 15% | 22% | |
| Body site | < 0.01* | ||
| blood | 8% | 20% | < 0.01 |
| screening at muco-cutaneous sites | 24% | 9% | < 0.01 |
| skin or soft tissue infections | 21% | 33% | 0.03 |
| respiratory tract | 24% | 22% | 0.71 |
| urines | 8% | 2% | 0.04 |
| other | 15% | 14% | 0.71 |
| . | Patients with MRSA . | Patients with MSSA . | P value . |
|---|---|---|---|
| Sex ratio (M/F) | 0.8 | 1.1 | 0.15* |
| Median age, years (range) | 78 (0–97) | 67 (1–96) | < 0.01** |
| Hospitalization units | 0.06* | ||
| medical wards | 32% | 28% | |
| intensive care units | 10% | 13% | |
| surgical wards | 18% | 22% | |
| geriatric wards | 26% | 15% | |
| other | 15% | 22% | |
| Body site | < 0.01* | ||
| blood | 8% | 20% | < 0.01 |
| screening at muco-cutaneous sites | 24% | 9% | < 0.01 |
| skin or soft tissue infections | 21% | 33% | 0.03 |
| respiratory tract | 24% | 22% | 0.71 |
| urines | 8% | 2% | 0.04 |
| other | 15% | 14% | 0.71 |
*χ2 test.
**Mann–Whitney test.
Sex, age, type of specimen and category of hospitalization unit of 510 patients with MRSA and 95 patients with MSSA infection or colonization
| . | Patients with MRSA . | Patients with MSSA . | P value . |
|---|---|---|---|
| Sex ratio (M/F) | 0.8 | 1.1 | 0.15* |
| Median age, years (range) | 78 (0–97) | 67 (1–96) | < 0.01** |
| Hospitalization units | 0.06* | ||
| medical wards | 32% | 28% | |
| intensive care units | 10% | 13% | |
| surgical wards | 18% | 22% | |
| geriatric wards | 26% | 15% | |
| other | 15% | 22% | |
| Body site | < 0.01* | ||
| blood | 8% | 20% | < 0.01 |
| screening at muco-cutaneous sites | 24% | 9% | < 0.01 |
| skin or soft tissue infections | 21% | 33% | 0.03 |
| respiratory tract | 24% | 22% | 0.71 |
| urines | 8% | 2% | 0.04 |
| other | 15% | 14% | 0.71 |
| . | Patients with MRSA . | Patients with MSSA . | P value . |
|---|---|---|---|
| Sex ratio (M/F) | 0.8 | 1.1 | 0.15* |
| Median age, years (range) | 78 (0–97) | 67 (1–96) | < 0.01** |
| Hospitalization units | 0.06* | ||
| medical wards | 32% | 28% | |
| intensive care units | 10% | 13% | |
| surgical wards | 18% | 22% | |
| geriatric wards | 26% | 15% | |
| other | 15% | 22% | |
| Body site | < 0.01* | ||
| blood | 8% | 20% | < 0.01 |
| screening at muco-cutaneous sites | 24% | 9% | < 0.01 |
| skin or soft tissue infections | 21% | 33% | 0.03 |
| respiratory tract | 24% | 22% | 0.71 |
| urines | 8% | 2% | 0.04 |
| other | 15% | 14% | 0.71 |
*χ2 test.
**Mann–Whitney test.
PFGE type distribution
The PFGE patterns of the 103 MSSA strains were classified into 24 groups and 40 types. Seventeen PFGE types, accounting for 79% of the strains, were classified as epidemic (Table 2). The 511 MRSA strains classified into 15 groups and 36 types, of which 9 epidemic PFGE types accounted for 90% of MRSA strains.17
Distribution of MSSA (n = 103) versus MRSA (n = 511) genotypes by PFGE/MLST-SCCmec and agr
| PFGE group . | PFGE type . | No. of MSSA isolates . | No. of MRSA isolatesa . | STa (no. of MSSA tested) . | CC . | SCCmeca (if MRSA) . | agra group (no. of MSSA tested) . | Corresponding international MRSA clones7,42 . |
|---|---|---|---|---|---|---|---|---|
| A | A1 | – | 12 | 247 | 8 | I | 1 | Iberian |
| A20 | 4 | 97 | 8 (1) | 8 | IV | 1 (1) | UK-EMRSA 2/6 – USA500 | |
| A21 | 3 | 27 | 8 (1) | 8 | IV | 1 (1) | ||
| other | 3 | 12 | ||||||
| B | B2 | 14 | 251 | 45 | 45 | IV | 1 | Berlin – USA600 |
| N (1) | 45 | 1 (1) | ||||||
| other | – | 18 | ||||||
| C | C1 | – | 13 | 5 | 5 | II | 2 | New York/Japan |
| 225 | 5 | |||||||
| C3 | 18 | 12 | 5 (1) | 5 | IV | 2 (1) | Pediatric-USA800 | |
| other | – | 9 | ||||||
| D | D8 | – | 11 | 228 | 5 | I | 2 | Southern Germany |
| G | G10 | – | 27 | 5 | 5 | II | 2 | New York/Japan |
| other | – | 4 | ||||||
| J | J2 | 7 | – | 642 (1) | 30 | – | 3 (1) | |
| J5 | 5 | – | 34 (1) | 30 | – | 3 (1) | ||
| J7 | 3 | – | 30 (1) | 30 | – | 3 (1) | South-West Pacific – USA1100 | |
| other | 2 | 1 | 30 | 30 | IV | 3 | ||
| L | L1 | 4 | 10 | 22 (1) | 22 | IV | 1 (1) | UK EMRSA 15 |
| other | – | 1 | ||||||
| O | O1 | 4 | – | 7 (1) | 7 | – | 1(1) | |
| O2 | 2 | – | 101 (1) | 101 | – | 1(1) | ||
| Q | Q3 | 2 | – | 12 (1) | 12 | – | 2(1) | |
| other | 2 | – | – | |||||
| R | R1 | 2 | – | 888 (1) | 9 | – | 2 (1) | |
| R2 | 2 | – | 109 (1) | 9 | – | 2 (1) | ||
| S | S1 | 2 | – | 1 (1) | 1 | – | 3 (1) | USA 400 (SCCmec IV) |
| other | 1 | – | – | |||||
| T | T1 | 3 | – | 15 (1) | 15 | – | 2 (1) | |
| other | 1 | – | – | |||||
| U | U1 | 2 | – | 25 (1) | 25 | – | 1 (1) | |
| U2 | 2 | – | 25 (1) | 25 | – | 1 (1) | ||
| P | 2 | – | 886 (1) | 30 | – | 3 (1) | ||
| N | 2 | – | 121 (2) | 121 | – | 4 (2) | ||
| V | 2 | – | 97 (1) | 97 | – | 1 (1) | ||
| Other | 9 | 7 |
| PFGE group . | PFGE type . | No. of MSSA isolates . | No. of MRSA isolatesa . | STa (no. of MSSA tested) . | CC . | SCCmeca (if MRSA) . | agra group (no. of MSSA tested) . | Corresponding international MRSA clones7,42 . |
|---|---|---|---|---|---|---|---|---|
| A | A1 | – | 12 | 247 | 8 | I | 1 | Iberian |
| A20 | 4 | 97 | 8 (1) | 8 | IV | 1 (1) | UK-EMRSA 2/6 – USA500 | |
| A21 | 3 | 27 | 8 (1) | 8 | IV | 1 (1) | ||
| other | 3 | 12 | ||||||
| B | B2 | 14 | 251 | 45 | 45 | IV | 1 | Berlin – USA600 |
| N (1) | 45 | 1 (1) | ||||||
| other | – | 18 | ||||||
| C | C1 | – | 13 | 5 | 5 | II | 2 | New York/Japan |
| 225 | 5 | |||||||
| C3 | 18 | 12 | 5 (1) | 5 | IV | 2 (1) | Pediatric-USA800 | |
| other | – | 9 | ||||||
| D | D8 | – | 11 | 228 | 5 | I | 2 | Southern Germany |
| G | G10 | – | 27 | 5 | 5 | II | 2 | New York/Japan |
| other | – | 4 | ||||||
| J | J2 | 7 | – | 642 (1) | 30 | – | 3 (1) | |
| J5 | 5 | – | 34 (1) | 30 | – | 3 (1) | ||
| J7 | 3 | – | 30 (1) | 30 | – | 3 (1) | South-West Pacific – USA1100 | |
| other | 2 | 1 | 30 | 30 | IV | 3 | ||
| L | L1 | 4 | 10 | 22 (1) | 22 | IV | 1 (1) | UK EMRSA 15 |
| other | – | 1 | ||||||
| O | O1 | 4 | – | 7 (1) | 7 | – | 1(1) | |
| O2 | 2 | – | 101 (1) | 101 | – | 1(1) | ||
| Q | Q3 | 2 | – | 12 (1) | 12 | – | 2(1) | |
| other | 2 | – | – | |||||
| R | R1 | 2 | – | 888 (1) | 9 | – | 2 (1) | |
| R2 | 2 | – | 109 (1) | 9 | – | 2 (1) | ||
| S | S1 | 2 | – | 1 (1) | 1 | – | 3 (1) | USA 400 (SCCmec IV) |
| other | 1 | – | – | |||||
| T | T1 | 3 | – | 15 (1) | 15 | – | 2 (1) | |
| other | 1 | – | – | |||||
| U | U1 | 2 | – | 25 (1) | 25 | – | 1 (1) | |
| U2 | 2 | – | 25 (1) | 25 | – | 1 (1) | ||
| P | 2 | – | 886 (1) | 30 | – | 3 (1) | ||
| N | 2 | – | 121 (2) | 121 | – | 4 (2) | ||
| V | 2 | – | 97 (1) | 97 | – | 1 (1) | ||
| Other | 9 | 7 |
N, new ST.
PFGE groups common to MRSA and MSSA are represented in bold font.
aMLST, SCCmec types and agr groups of MRSA are from ref. 17.
Distribution of MSSA (n = 103) versus MRSA (n = 511) genotypes by PFGE/MLST-SCCmec and agr
| PFGE group . | PFGE type . | No. of MSSA isolates . | No. of MRSA isolatesa . | STa (no. of MSSA tested) . | CC . | SCCmeca (if MRSA) . | agra group (no. of MSSA tested) . | Corresponding international MRSA clones7,42 . |
|---|---|---|---|---|---|---|---|---|
| A | A1 | – | 12 | 247 | 8 | I | 1 | Iberian |
| A20 | 4 | 97 | 8 (1) | 8 | IV | 1 (1) | UK-EMRSA 2/6 – USA500 | |
| A21 | 3 | 27 | 8 (1) | 8 | IV | 1 (1) | ||
| other | 3 | 12 | ||||||
| B | B2 | 14 | 251 | 45 | 45 | IV | 1 | Berlin – USA600 |
| N (1) | 45 | 1 (1) | ||||||
| other | – | 18 | ||||||
| C | C1 | – | 13 | 5 | 5 | II | 2 | New York/Japan |
| 225 | 5 | |||||||
| C3 | 18 | 12 | 5 (1) | 5 | IV | 2 (1) | Pediatric-USA800 | |
| other | – | 9 | ||||||
| D | D8 | – | 11 | 228 | 5 | I | 2 | Southern Germany |
| G | G10 | – | 27 | 5 | 5 | II | 2 | New York/Japan |
| other | – | 4 | ||||||
| J | J2 | 7 | – | 642 (1) | 30 | – | 3 (1) | |
| J5 | 5 | – | 34 (1) | 30 | – | 3 (1) | ||
| J7 | 3 | – | 30 (1) | 30 | – | 3 (1) | South-West Pacific – USA1100 | |
| other | 2 | 1 | 30 | 30 | IV | 3 | ||
| L | L1 | 4 | 10 | 22 (1) | 22 | IV | 1 (1) | UK EMRSA 15 |
| other | – | 1 | ||||||
| O | O1 | 4 | – | 7 (1) | 7 | – | 1(1) | |
| O2 | 2 | – | 101 (1) | 101 | – | 1(1) | ||
| Q | Q3 | 2 | – | 12 (1) | 12 | – | 2(1) | |
| other | 2 | – | – | |||||
| R | R1 | 2 | – | 888 (1) | 9 | – | 2 (1) | |
| R2 | 2 | – | 109 (1) | 9 | – | 2 (1) | ||
| S | S1 | 2 | – | 1 (1) | 1 | – | 3 (1) | USA 400 (SCCmec IV) |
| other | 1 | – | – | |||||
| T | T1 | 3 | – | 15 (1) | 15 | – | 2 (1) | |
| other | 1 | – | – | |||||
| U | U1 | 2 | – | 25 (1) | 25 | – | 1 (1) | |
| U2 | 2 | – | 25 (1) | 25 | – | 1 (1) | ||
| P | 2 | – | 886 (1) | 30 | – | 3 (1) | ||
| N | 2 | – | 121 (2) | 121 | – | 4 (2) | ||
| V | 2 | – | 97 (1) | 97 | – | 1 (1) | ||
| Other | 9 | 7 |
| PFGE group . | PFGE type . | No. of MSSA isolates . | No. of MRSA isolatesa . | STa (no. of MSSA tested) . | CC . | SCCmeca (if MRSA) . | agra group (no. of MSSA tested) . | Corresponding international MRSA clones7,42 . |
|---|---|---|---|---|---|---|---|---|
| A | A1 | – | 12 | 247 | 8 | I | 1 | Iberian |
| A20 | 4 | 97 | 8 (1) | 8 | IV | 1 (1) | UK-EMRSA 2/6 – USA500 | |
| A21 | 3 | 27 | 8 (1) | 8 | IV | 1 (1) | ||
| other | 3 | 12 | ||||||
| B | B2 | 14 | 251 | 45 | 45 | IV | 1 | Berlin – USA600 |
| N (1) | 45 | 1 (1) | ||||||
| other | – | 18 | ||||||
| C | C1 | – | 13 | 5 | 5 | II | 2 | New York/Japan |
| 225 | 5 | |||||||
| C3 | 18 | 12 | 5 (1) | 5 | IV | 2 (1) | Pediatric-USA800 | |
| other | – | 9 | ||||||
| D | D8 | – | 11 | 228 | 5 | I | 2 | Southern Germany |
| G | G10 | – | 27 | 5 | 5 | II | 2 | New York/Japan |
| other | – | 4 | ||||||
| J | J2 | 7 | – | 642 (1) | 30 | – | 3 (1) | |
| J5 | 5 | – | 34 (1) | 30 | – | 3 (1) | ||
| J7 | 3 | – | 30 (1) | 30 | – | 3 (1) | South-West Pacific – USA1100 | |
| other | 2 | 1 | 30 | 30 | IV | 3 | ||
| L | L1 | 4 | 10 | 22 (1) | 22 | IV | 1 (1) | UK EMRSA 15 |
| other | – | 1 | ||||||
| O | O1 | 4 | – | 7 (1) | 7 | – | 1(1) | |
| O2 | 2 | – | 101 (1) | 101 | – | 1(1) | ||
| Q | Q3 | 2 | – | 12 (1) | 12 | – | 2(1) | |
| other | 2 | – | – | |||||
| R | R1 | 2 | – | 888 (1) | 9 | – | 2 (1) | |
| R2 | 2 | – | 109 (1) | 9 | – | 2 (1) | ||
| S | S1 | 2 | – | 1 (1) | 1 | – | 3 (1) | USA 400 (SCCmec IV) |
| other | 1 | – | – | |||||
| T | T1 | 3 | – | 15 (1) | 15 | – | 2 (1) | |
| other | 1 | – | – | |||||
| U | U1 | 2 | – | 25 (1) | 25 | – | 1 (1) | |
| U2 | 2 | – | 25 (1) | 25 | – | 1 (1) | ||
| P | 2 | – | 886 (1) | 30 | – | 3 (1) | ||
| N | 2 | – | 121 (2) | 121 | – | 4 (2) | ||
| V | 2 | – | 97 (1) | 97 | – | 1 (1) | ||
| Other | 9 | 7 |
N, new ST.
PFGE groups common to MRSA and MSSA are represented in bold font.
aMLST, SCCmec types and agr groups of MRSA are from ref. 17.
Despite the larger diversity in their PFGE pattern distributions, 45% of MSSA strains clustered in the same five epidemic PFGE types as 76% of MRSA strains: PFGE types C3 (17% of MSSA strains versus 2% of MRSA strains), B2 (14% versus 49%), A20 (5% versus 18%), A21 (3% versus 5%) and L1 (4% versus 2%) (Table 2 and Figure 1). Strikingly, all MRSA strains belonging to those types harboured the type IV SCCmec element. Seventeen MSSA strains (17%) and a single MRSA strain clustered in the PFGE group J.
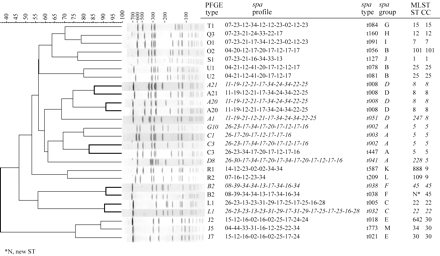
Dendrogram of PFGE profiles of representative major epidemic MRSA (italics) and MSSA. For each isolate represented, spa profile, spa type, spa group, MLST ST and CC are listed. Branches linking the MSSA and MRSA profiles belonging to the same PFGE type are thickened.
spa type distribution
spa typing was performed on representative MSSA (n = 17) and MRSA (n = 9) strains of each epidemic PFGE type. Twenty-one spa types were found, distributed in five groups and eight singletons (groups represented by a single type). spa typing confirmed PFGE analysis by clustering together MSSA and MRSA strains with common PFGE type within the same spa groups (Figure 1). These groups were clearly distinct from those that included MSSA epidemic types without MRSA homologue. PFGE types B2, A20 and A21 MSSA strains shared exactly the same spa type as their MRSA homologues. PFGE types C3 and L1 MRSA strains differed in spa type from their MSSA homologues by addition of one and four repeats, respectively. However, spa typing diverged from PFGE by grouping strains belonging to distinct PFGE groups (e.g. G10, C3, C1 and D8) in the same spa group. Furthermore, the G10 and C3 MRSA strains shared the same spa type.
MSSA genetic background
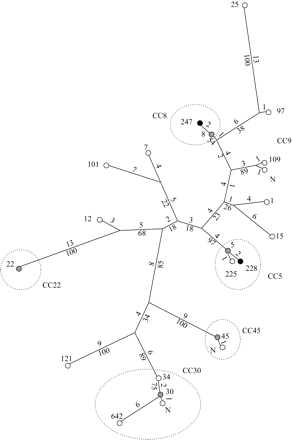
To compare the genetic backgrounds of Belgian major epidemic MSSA clones with international epidemiological data, MLST was performed on one strain of each PFGE group. The 20 strains tested were classified into 18 STs and 15 CCs (Table 2). The MSSA strains belonging to the epidemic PFGE types A20-21, C3 and L1 MSSA strains clustered in the same STs as their MRSA homologues: ST8, ST5 and ST22 respectively. The type B2 MSSA strain was a single locus variant (SLV) (one point mutation on the tpi fragment) of its ST45-SCCmec IV MRSA homologue. Strains belonging to the PFGE group J clustered along with the PFGE group P strain in CC30 but belonged to different STs (Table 2). PFGE group P isolate harboured ST886, an SLV of ST30 (one point mutation on the aroE fragment). All together, 63% of MSSA isolates belonged to one of the five major CC of international MRSA pandemic clones (CC30, CC5, CC45, CC8 and CC22, Figure 2).
Maximum parsimony tree of representative major epidemic MRSA and MSSA based on the concatenated sequences of the seven fragments of housekeeping genes used for MLST. The absolute numbers of nucleotide differences between STs are indicated above the branches, the bootstrap values below. ST represented only by MSSA isolates are represented by white circles; ST represented only by MRSA isolates are represented by black circles; and ST represented by both MRSA and MSSA isolates are represented by grey circles. Dotted line circles delineate the five major MRSA CCs.
The strains representing the nine epidemic MSSA-PFGE types without MRSA homologues showed very diverse MLST profiles (Table 2), confirming the large diversity found by the PFGE typing. They clustered in eight different STs, which were all phylogenetically distant from the five major international MRSA CCs (Figure 2).
Toxin genes detection and agr group distribution
The majority of strains were negative for both PVL and TSST-1 genes. Among MSSA strains, 12 were TSST-1 positive and 3 were PVL positive. One strain carried the two toxins. Strikingly, all the toxin-harbouring MSSA strains belonged to CC30 and were clustered in either PFGE group P or PFGE group J. Among the 156 representative MRSA strains tested, 14 harboured TSST-1 gene (8 belonging to PFGE type G10, 2 to L1 and one to A21, B2 and C1 each). One PFGE J strain carried the PVL genes.17
The agr group 1 was the most frequent agr group among both MSSA and MRSA strains (43% and 85% respectively, Table 2) followed by group 2 (33% of MSSA strains and 38% of MRSA strains). Group 3 was found in 22% of MSSA strains and in only one (0.2%) MRSA strain, all belonging to CC30. Only two MSSA strains, both of ST121, were agr group 4.
Discussion
The present study is, to our knowledge, the first cross-sectional comparison of the genetic background of MSSA and MRSA strains prospectively and randomly selected at a national scale. Ideally, the best sampling strategy to test the hypothesis of MRSA clones arising locally via SCCmec IV acquisition by successful MSSA clones would have been to retrieve a nationally representative sample of MSSA isolates from a time period preceding the emergence of these MRSA clones. As such a sample was not available in our collection, we opted for a cross-sectional survey by parallel sampling of MSSA and MRSA from hospitalized patients across the country.
Due to the study design, MRSA strains were more frequently isolated from screening swabs than the MSSA strains, inducing a selection bias regarding the colonization/infection ratio in the two groups. Patients carrying MRSA strains were also older than patients carrying MSSA strains, which was predictable, since advanced age is known to be a risk factor for carrying MRSA.25,26
Despite the wide genotypic diversity found among the MSSA strains, 45% clustered in the same five PFGE epidemic types as 76% of MRSA strains. Those five HA-MRSA epidemic clones all harboured type IV SCCmec and have emerged after 1994 in Belgian hospitals. They also correspond by MLST to well known pandemic MRSA clones, namely the UK-EMRSA 2/6 and 15, Pediatric and Berlin clones. Overall, spa typing confirmed the PFGE analysis by clustering together MSSA and MRSA strains with common PFGE type within the same spa groups. However, minor discrepancies were noted between spa typing and PFGE analysis at type level. The PFGE types C3 and L1 MRSA strains differed from their MSSA homologues by addition of 1 and 4 repeats, respectively. Conversely, PFGE types G10 and C3 MRSA strains (both belonging to ST5) shared the same spa type. Those discrepancies between spa and PFGE, can, at least partially, be explained if the target of the typing method is taken into account. Indeed, the PFGE method, which is based on the restriction of the whole genome, indexes variations in the presence of mobile elements. One can speculate that insertion of SCCmec of different type and size (type II for the G10 PFGE type and type IV for the C3 PFGE type strain) in strains of otherwise similar genetic background, leads to differences between their PFGE patterns while their spa types remain identical.
The identification of common genetic backgrounds between successful MSSA clones and epidemic HA-MRSA clones suggests that these genetic backgrounds could confer a selective advantage, regardless of the resistance profile. The fact that those MRSA epidemic clones all emerged recently and harbour type IV SCCmec argues in favour of the frequent and recent acquisition of the more mobile SCCmec type IV. However, when compared to studies conducted in other countries or at an international scale,7 the predominant MSSA clones identified in this study (ST8, ST45, ST5, ST30 and ST34) were also frequently found in other countries, indicating the presence of pandemic MSSA lineages.23,27 Since those MSSA clones as well as their MRSA homologues are disseminated worldwide and repeated acquisition of the same SCCmec type by the same ST cannot be discerned,7 no conclusion can be drawn so far regarding the frequency and geographic origin of SCCmec IV acquisition. Another possibility that cannot be excluded, is that a fraction of the MSSA strains sharing similar genotype with MRSA could be derived from MRSA strains by the loss of the mecA gene.28,29 However, the risk of an in vitro mecA loss is low since these strains were collected and identified as MSSA directly from the clinical sample and prior to any frozen storage.
Up to 31% of MSSA strains examined here belonged to nine CCs which are distinct from the five major international MRSA CCs. Besides PVL-positive ST1-SCCmec IV CA-MRSA described in the USA,30 few or no MRSA strains have been, to our knowledge, described within those CCs. This may indicate that the fact of being successful is not, by itself, sufficient for the acquisition of SCCmec, which supports the hypothesis that some genetic backgrounds may be more ‘receptive’ to SCCmec than others.31 We cannot rule out the hypothesis that some successful MRSA lineages could arise in the future from those MSSA clones. Such a sequential wave phenomenon has already been described for early MRSA strains in Denmark.32
A significant proportion of MSSA isolates clustered in CC30, presenting the same PFGE group (J) as some community-acquired PVL-positive MRSA strains recently found in Belgium,15 belonging to the ST30-SCCmec IV genotype (South-West Pacific clone) and closely related to EMRSA 16, which has recently spread in Europe.33 Seventy-three percent of these strains carried TSST-1 or PVL genes, known to be potent virulence genes. In fact, all TSST-1 or PVL-positive MSSA strains belonged to CC30 (ST30, 642 and 34). This is consistent with the findings of other investigators that TSST-1 is highly clonal and found in association with the agr 3 CC30 or CC39 lineages.34,35 Peacock et al.35 attribute this phenomenon to a recent and highly successful clonal expansion, leaving less time for those CCs to diversify through horizontal transfer. Strikingly, no Belgian HA-MRSA epidemic clone belongs to this CC30. Again, we cannot exclude that, in the future, clones combining resistance and virulence could arise via the local introduction of SCCmec in this successful lineage. This could especially be true if the expansion of this lineage is relatively recent among MSSA, as suggested by Peacock et al.35
TSST-1 was also found among Belgian MRSA. Its distribution seems, by contrast, far more divergent: MRSA isolates belonging to various genetic backgrounds (ST8-SCCmec IV, ST5-SCCmec II, ST225-SCCmec IV, ST22-SCCmec IV and ST5-SCCmec II) were found to carry TSST-1.17 A large majority of them (72%) belonged to the PFGE G10 ST5-SCCmec II (New-York/Japan clone36), which has been associated with well-documented toxic shock syndrome.36 However, although strongly lineage-associated, TSST-1 is known to be located on a mobile genetic element,37 and thus can be horizontally transferred. This could explain why TSST-1 was found in isolates belonging to several successful MRSA lineages.
Like other countries, Belgium is facing the emergence of PVL positive CA-MRSA.15 Four major lineages were described in our country so far: ST80-SCCmec IV, ST30-SCCmec IV, ST88-SCCmec IV and ST8-SCCmec IV. Except for ST8-SCCmec IV strains that appeared closely related to PFGE A20/A21 HA-MRSA, the majority of strains (ST80, ST30 and ST88) had a genetic background that is clearly distinct from those of HA-MRSA, suggesting importation of these strains.15 In the present study, we identified ST8 as well as ST30 epidemic MSSA (some of which carry PVL genes), a finding which could favour local emergence as a likely scenario for these clones. However, no ST80 MSSA were found, indicating that ST80-SCCmec IV CA-MRSA, the predominant clone in Belgium, as in other European countries,15,38 was probably imported from abroad.
The determination of agr groups, performed on representative strains of major epidemic types showed that agr group 1 was the most frequent type among both MRSA and MSSA, followed by agr group 2. In contrast, agr group 3 (harboured by CC30 strains) was frequently recovered among MSSA in association with toxin carriage but rarely found among HA-MRSA. Two ST121 MSSA strains were agr group 4. These are the two first human isolates identified in our country belonging to this agr group. Interestingly, outbreaks due to a highly virulent ST121-agr 4 MSSA have recently been described in Belgian rabbitries.39
In conclusion, we present here the first cross-sectional study of the genetic population structure of MSSA strains at a national scale in Belgium. The identification of common genetic backgrounds of successful MSSA and SCCmec IV-harbouring MRSA clones is consistent with the frequent and recent acquisition of the more mobile type IV SCCmec element by MSSA. This finding also raises the issue of genetic backgrounds conferring a selective advantage to S. aureus, regardless of the antibiotic-resistance profile. However, since the MSSA and MRSA lineages present in Belgium seem to be disseminated world-wide and multiple acquisitions of SCCmec IV type into the same ST cannot be discerned, no conclusion can be made regarding the place and time of acquisition of SCCmec type IV in locally and globally successful MSSA strains as source of epidemic MRSA strains in Belgium. High-resolution typing methods such as MLST extended to S. aureus surface protein (sas) genes,7 combined with extensive characterization of variants within SCCmec type IV,40 could help resolve this question. Since frequent horizontal transfer of type IV SCCmec and other mobile genetic elements seem to be the source of the ongoing genetic diversification of MRSA,7,31,41 large MSSA population genetic studies, as the one reported here, should be conducted in parallel with MRSA surveillance studies, to understand how natural populations of MS- and MRSA co-evolve and interact.
Acknowledgements
This work was supported by a grant from the Fonds Erasme, Brussels, Belgium.
Transparency declarations
None to declare.