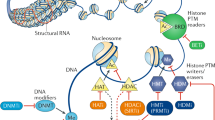
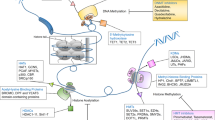
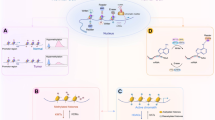
Abstract
The promise of targeting epigenetic abnormalities for cancer therapy has not been realized for solid tumours, although increasing evidence is demonstrating its worth in haematological malignancies. In fact, true clinical efficacy in haematopoietic-related neoplasms has only become evident at low doses of epigenetic-targeting drugs (namely, inhibitors of histone deacetylase and DNA methyltransferases). Describing data from preclinical studies and early clinical trial results, we hypothesize that in using low-dose epigenetic-modulating agents, tumour cells can be reprogrammed, which overrides any immediate cytotoxic and off-target effect observed at high dose. We suggest that such optimization of drug dosing and scheduling of currently available agents could give these agents a prominent place in cancer management—when used alone or in combination with other therapies. If so, optimal use of these known agents might also pave the way for the introduction of other agents that target the epigenome.
Key Points
-
Evidence of the effects of epigenetic-modulating agents has revealed their dramatic consequences on cellular programming, in particular reversing stem-cell-like behaviour and chemoresistance
-
Treatment with epigenetic drugs affects multiple cell signalling pathways, including those regulating immune response and evasion, apoptosis, cell survival and DNA-damage repair
-
If used optimally, the widespread targets of these agents can be their greatest feature—cancer cells abnormally regulate many diverse pathways, which likely results in major therapeutic barriers
-
Previous research and clinical use of these agents might have been hampered because their effects were assessed too early and the doses used were too high
-
Lower doses of these agents results in less cytotoxicity to normal tissue, and should provide the added time and exposure needed for the 'reprogramming' effect of epigenetic therapy to become evident
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 (2002).
Jones, P. A. & Baylin, S. B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Ecker, J. R. et al. Genomics: ENCODE explained. Nature 489, 52–55 (2012).
Holliday, R. DNA methylation and epigenetic mechanisms. Cell Biophys. 15, 15–20 (1989).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Caparros, M. (Ed.) Epigenetics (Cold Spring Harbor Laboratory Press, 2007).
Allis, C. D., Jenuwein, T. & Reinberg, D. (Eds) Epigenetics (Cold Spring Harbor Laboratory Press, 2007).
van der Vlag, J. & Otte, A. P. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23, 474–478 (1999).
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010).
Thol, F. et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J. Clin. Oncol. 29, 2889–2896 (2011).
Metzeler, K. H. et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 29, 1373–1381 (2011).
Bejar, R. et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364, 2496–2506 (2011).
Abdel-Wahab, O. et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia 25, 1200–1202 (2011).
Metzeler, K. H. et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN favorable genetic category. Blood 118, 6920–6929 (2011).
Guglielmelli, P. et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood 118, 5227–5234 (2011).
Tefferi, A. et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23, 905–911 (2009).
Tefferi, A. et al. IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia 26, 475–480 (2012).
Tefferi, A. et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 24, 1302–1309 (2010).
De Carvalho, D. D. et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 21, 655–667 (2012).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Berdasco, M. & Esteller, M. Genetic syndromes caused by mutations in epigenetic genes. Hum. Genet. 132, 359–383 (2013).
Shih, A. H., Abdel-Wahab, O., Patel, J. P. & Levine, R. L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 12, 599–612 (2012).
Kaminskas, E. et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin. Cancer Res. 11, 3604–3608 (2005).
Silverman, L. R. et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 20, 2429–2440 (2002).
Kaminskas, E., Farrell, A. T., Wang, Y. C., Sridhara, R. & Pazdur, R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 10, 176–182 (2005).
Kantarjian, H. et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 106, 1794–1803 (2006).
Piekarz, R. L. et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J. Clin. Oncol. 27, 5410–5417 (2009).
Whittaker, S. J. et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 28, 4485–4491 (2010).
O'Connor, O. A. et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J. Clin. Oncol. 24, 166–173 (2006).
Duvic, M. et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109, 31–39 (2007).
Issa, J. P. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat. Clin. Pract. Oncol. 2 (Suppl. 1), S24–29 (2005).
Herman, J. G. & Baylin, S. B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 349, 2042–2054 (2003).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Doi, A. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353 (2009).
Lane, A. A. & Chabner, B. A. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 27, 5459–5468 (2009).
Popovic, R. & Licht, J. D. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2, 405–413 (2012).
Dawson, M. A. & Kouzarides, T. Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012).
Ohm, J. E. et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 (2007).
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Issa, J. P. & Kantarjian, H. M. Introduction: emerging role of epigenetic therapy: focus on decitabine. Semin. Hematol. 42 (Suppl.), S1–S2 (2005).
Egger, G., Liang, G., Aparicio, A. & Jones, P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004).
Hochedlinger, K. & Plath, K. Epigenetic reprogramming and induced pluripotency. Development 136, 509–523 (2009).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Feng, B., Ng, J. H., Heng, J. C. & Ng, H. H. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4, 301–312 (2009).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Morey, L. et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62 (2012).
Easwaran, H. et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22, 837–849 (2012).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Tsao, M. S. et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N. Engl. J. Med. 353, 133–144 (2005).
Nahta, R., Yu, D., Hung., M. C., Hortobagyi, G. N. & Esteva, F. J. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat. Clin. Pract. Oncol. 3, 269–280 (2006).
Astsaturov, I. et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci. Signal. 3, ra67 (2010).
Poulikakos, P. I. & Rosen, N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell 19, 11–15 (2011).
Solit, D. B. & Rosen, N. Resistance to BRAF inhibition in melanomas. N. Engl. J. Med. 364, 772–774 (2011).
Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007).
Sorm, F. & Vesely, J. Effect of 5-aza-2'-deoxycytidine against leukemic and hemopoietic tissues in AKR mice. Neoplasma 15, 339–343 (1968).
Notari, R. E. & DeYoung, J. L. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J. Pharm. Sci. 64, 1148–1157 (1975).
Jones, P. A. & Taylor, S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980).
Juergens, R. A. et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 1, 598–607 (2011).
Ferguson, A. T. et al. Role of estrogen receptor gene demethylation and DNA methyltransferase.DNA adduct formation in 5-aza-2'deoxycytidine-induced cytotoxicity in human breast cancer cells. J. Biol. Chem. 272, 32260–32266 (1997).
Gabbara, S. & Bhagwat, A. S. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem. J. 307, 87–92 (1995).
Santi, D. V., Norment, A. & Garrett, C. E. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl Acad. Sci. USA 81, 6993–6997 (1984).
Ghoshal, K. et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell Biol. 25, 4727–4741 (2005).
Tsai, H. C. et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21, 430–446 (2012).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Silverman, L. R. et al. Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer 117, 2697–2702 (2011).
Fandy, T. E. et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood 114, 2764–2773 (2009).
Brock, M. V. et al. DNA methylation markers and early recurrence in stage I lung cancer. N. Engl. J. Med. 358, 1118–1128 (2008).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Hammerman, P. S. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Koboldt, D. C. et al. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Saito, Y. et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443 (2006).
You, J. S. & Jones, P. A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 (2012).
Noushmehr, H. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Suter, C. M., Martin, D. I. & Ward, R. L. Germline epimutation of MLH1 in individuals with multiple cancers. Nat. Genet. 36, 497–501 (2004).
Chan, E. C., Lam, S. Y., Fu, K. H. & Kwong, Y. L. Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: relationship with aberrant promoter methylation of the CDKN2A and RARB genes. Cancer Genet. Cytogenet. 162, 10–20 (2005).
Chan, T. L. et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat. Genet. 38, 1178–1183 (2006).
Ligtenberg, M. J. et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat. Genet. 41, 112–117 (2009).
Girard, N. et al. Third-line chemotherapy in advanced non-small cell lung cancer: identifying the candidates for routine practice. J. Thorac. Oncol. 4, 1544–1549 (2009).
Humeniuk, R. et al. Decreased levels of UMP kinase as a mechanism of fluoropyrimidine resistance. Mol. Cancer Ther. 8, 1037–1044 (2009).
Meng, F., Sun, G., Zhong, M., Yu, Y. & Brewer, M. A. Anticancer efficacy of cisplatin and trichostatin A or 5-aza-2'-deoxycytidine on ovarian cancer. Br. J. Cancer 108, 579–586 (2013).
Bhatla, T. et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood 119, 5201–5210 (2012).
Shakya, R. et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res. 73, 885–896 (2012).
Fu, S. et al. Phase 1b–2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer 117, 1661–1669 (2011).
Matei, D. et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 72, 2197–2205 (2012).
Gray, S. G. & Ekström, T. J. The human histone deacetylase family. Exp. Cell Res. 262, 75–83 (2001).
Solomon, J. M. et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell Biol. 26, 28–38 (2006).
O'Hagan, H. M., Mohammad, H. P. & Baylin, S. B. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 4, e1000155 (2008).
Vaquero, A. et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256–1261 (2006).
Pruitt, K. et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2, e40 (2006).
Pediconi, N. et al. hSirT1-dependent regulation of the PCAF–E2F1–p73 apoptotic pathway in response to DNA damage. Mol. Cell Biol. 29, 1989–1998 (2009).
Cooper, S. J. et al. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol. Cancer Ther. 11, 2105–2115 (2012).
Isozaki, Y. et al. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int. J. Oncol. 41, 985–994 (2012).
Shin, H., Lee, Y. S. & Lee, Y. C. Sodium butyrate-induced DAPK-mediated apoptosis in human gastric cancer cells. Oncol. Rep. 27, 1111–1115 (2012).
Mund, C. & Lyko, F. Epigenetic cancer therapy: Proof of concept and remaining challenges. Bioessays 32, 949–957 (2010).
Verbrugge, I., Johnstone, R. W. & Bots, M. Promises and challenges of anticancer drugs that target the epigenome. Epigenomics 3, 547–565 (2011).
Robert, C. & Rassool, F. V. HDAC inhibitors: roles of DNA damage and repair. Adv. Cancer Res. 116, 87–129 (2012).
Kachhap, S. K. et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS ONE 5, e11208 (2010).
Gilbertson, R. J. & Graham, T. A. Cancer: resolving the stem-cell debate. Nature 488, 462–463 (2012).
Chen, J. et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526 (2012).
Driessens, G., Beck, B., Caauwe, A., Simons, B. D. & Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530 (2012).
Schepers, A. G. et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012).
Villanueva, J. et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18, 683–695 (2010).
Yeo, W. et al. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J. Clin. Oncol. 30, 3361–3367 (2012).
Hainsworth, J. D. et al. A phase II trial of panobinostat, a histone deacetylase inhibitor, in the treatment of patients with refractory metastatic renal cell carcinoma. Cancer Invest. 29, 451–455 (2011).
Giaccone, G. et al. Phase II study of belinostat in patients with recurrent or refractory advanced thymic epithelial tumors. J. Clin. Oncol. 29, 2052–2059 (2011).
Iwamoto, F. M. et al. A phase I/II trial of the histone deacetylase inhibitor romidepsin for adults with recurrent malignant glioma: North American Brain Tumor Consortium Study 03–03. Neuro. Oncol. 13, 509–516 (2011).
Mackay, H. J. et al. Phase II trial of the histone deacetylase inhibitor belinostat in women with platinum resistant epithelial ovarian cancer and micropapillary (LMP) ovarian tumours. Eur. J. Cancer 46, 1573–1579 (2010).
Witta, S. E. et al. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J. Clin. Oncol. 30, 2248–2255 (2012).
Ramalingam, S. S. et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J. Clin. Oncol. 28, 56–62 (2010).
Yardley, D. A., Ismail-Khan, R. & Klein, P. Results of ENCORE 301, a randomized, phase II, double-blind, placebo-controlled study of exemestane with or without entinostat in postmenopausal women with locally recurrent or metastatic estrogen receptor-positive (ER+) breast cancer progressing on a nonsteroidal aromatase inhibitor (AI) [abstract 268]. J. Clin. Oncol. 29 (Suppl. 27), 268 (2011).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Topalian, S. L. et al. Safety, activity, and immune orrelates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Issa, J. P. et al. Interim results from a randomized phase 1–2 first-in-human (FIH) study of PK/PD guided escalating doses of SGI-110, a novel subcutaneous (SQ) second generation hypomethylating agent (HMA) in relapsed/refractory MDS and AML [abstract LB-214]. Cancer Res. 72 (Suppl. 1), LB-214 (2012).
Garcia-Manero, G. et al. Phase I study of oral azacitidine in myelodysplastic syndromes, chronic myelomonocytic leukemia, and acute myeloid leukemia. J. Clin. Oncol. 29, 2521–2527 (2011).
Chung, C. W. et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem. 54, 3827–3838 (2011).
Spannhoff, A., Hauser, A. T., Heinke, R., Sippl, W. & Jung, M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4, 1568–1582 (2009).
Bernt, K. M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011).
Daigle, S. R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Olsen, E. A. et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 25, 3109–3115 (2007).
Ooi, S. K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
Hashimoto, H., Vertino, P. M. & Cheng, X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2, 657–669 (2010).
Kantarjian, H. et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood 109, 52–57 (2007).
Acknowledgements
A portion of the authors' work cited in this article was supported by grants from SU2C (Stand Up To Cancer), Waxman Foundation, Hodson Endowment and the NCI (National Cancer Institute) CA043318. The authors thank Kathy Bender for help with manuscript preparation and submission.
Author information
Authors and Affiliations
Contributions
N. Azad and S. B. Baylin researched the data for the article and wrote the manuscript. All authors contributed to the discussion of the article's content. C. A. Zahnow and C. M. Rudin edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
S. B. Baylin acts as a consultant for MDxHealth and is entitled (with Johns Hopkins University) to received royalty shares from the sales of the methylation-specific PCR (MSP) assay.
Rights and permissions
About this article
Cite this article
Azad, N., Zahnow, C., Rudin, C. et al. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol 10, 256–266 (2013). https://doi.org/10.1038/nrclinonc.2013.42
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2013.42
This article is cited by
-
Clinical advances in epigenetic therapies for lymphoma
Clinical Epigenetics (2023)
-
Functional characterization of age-dependent p16 epimutation reveals biological drivers and therapeutic targets for colorectal cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
Gene body methylation in cancer: molecular mechanisms and clinical applications
Clinical Epigenetics (2022)
-
Laboratory methods to decipher epigenetic signatures: a comparative review
Cellular & Molecular Biology Letters (2021)
-
HDAC inhibition potentiates anti-tumor activity of macrophages and enhances anti-PD-L1-mediated tumor suppression
Oncogene (2021)