Abstract
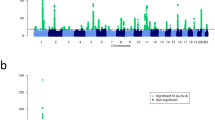
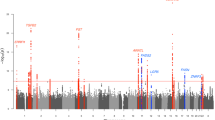
Atopic dermatitis is a chronic, relapsing form of inflammatory skin disorder that is affected by genetic and environmental factors. We performed a genome-wide association study of atopic dermatitis in a Chinese Han population using 1,012 affected individuals (cases) and 1,362 controls followed by a replication study in an additional 3,624 cases and 12,197 controls of Chinese Han ethnicity, as well as 1,806 cases and 3,256 controls from Germany. We identified previously undescribed susceptibility loci at 5q22.1 (TMEM232 and SLC25A46, rs7701890, Pcombined = 3.15 × 10−9, odds ratio (OR) = 1.24) and 20q13.33 (TNFRSF6B and ZGPAT, rs6010620, Pcombined = 3.0 × 10−8, OR = 1.17) and replicated another previously reported locus at 1q21.3 (FLG, rs3126085, Pcombined = 5.90 × 10−12, OR = 0.82) in the Chinese sample. The 20q13.33 locus also showed evidence for association in the German sample (rs6010620, P = 2.87 × 10−5, OR = 1.25). Our study identifies new genetic susceptibility factors and suggests previously unidentified biological pathways in atopic dermatitis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Williams, H., Stewart, A., von Mutius, E., Cookson, W. & Anderson, H.R. Is eczema really on the increase worldwide? J. Allergy Clin. Immunol. 121, 947–954 (2008).
Bieber, T. & Novak, N. Pathogenesis of atopic dermatitis: new developments. Curr. Allergy Asthma Rep. 9, 291–294 (2009).
Barnes, K.C. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J. Allergy Clin. Immunol. 125, 16–29 (2010).
Esparza-Gordillo, J. et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 41, 596–601 (2009).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Ioannidis, J.P., Ntzani, E.E., Trikalinos, T.A. & Contopoulos-Ioannidis, D.G. Replication validity of genetic association studies. Nat. Genet. 29, 306–309 (2001).
Zhang, X.J. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
Quan, C. et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat. Genet. 42, 614–618 (2010).
Han, J.W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Zhang, F.R. et al. Genomewide association study of leprosy. N. Engl. J. Med. 361, 2609–2618 (2009).
Palmieri, F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 (2004).
Wan, X., Shi, G., Semenuk, M., Zhang, J. & Wu, J. DcR3/TR6 modulates immune cell interactions. J. Cell. Biochem. 89, 603–612 (2003).
Zhang, J. et al. Modulation of T-cell responses to alloantigens by TR6/DcR3. J. Clin. In vest. 107, 1459–1468 (2001).
Hsu, T.L. et al. Modulation of dendritic cell differentiation and maturation by decoy receptor 3. J. Immunol. 168, 4846–4853 (2002).
You, R.I. et al. Apoptosis of dendritic cells induced by decoy receptor 3 (DcR3). Blood 111, 1480–1488 (2008).
Chang, Y.C. et al. Modulation of macrophage differentiation and activation by decoy receptor 3. J. Leukoc. Biol. 75, 486–494 (2004).
Chen, C.C., Yang, Y.H., Lin, Y.T., Hsieh, S.L. & Chiang, B.L. Soluble decoy receptor 3: increased levels in atopic patients. J. Allergy Clin. Immunol. 114, 195–197 (2004).
Li, R. et al. ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 28, 2763–2776 (2009).
Mascia, F., Mariani, V., Girolomoni, G. & Pastore, S. Blockade of the EGF receptor induces a deranged chemokine expression in keratinocytes leading to enhanced skin inflammation. Am. J. Pathol. 163, 303–312 (2003).
Wang, Q. et al. Characterization of Su48, a centrosome protein essential for cell division. Proc. Natl. Acad. Sci. USA 103, 6512–6517 (2006).
Gudbjartsson, D.F. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 41, 342–347 (2009).
Szczepankiewicz, A., Breborowicz, A., Sobkowiak, P., Kramer, L. & Popiel, A. Role of ADRB2 gene polymorphism in asthma and response to beta(2)-agonists in Polish children. J. Appl. Genet. 50, 275–281 (2009).
Tsunemi, Y. et al. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 30, 161–166 (2002).
Randolph, A.G. et al. The IL12B gene is associated with asthma. Am. J. Hum. Genet. 75, 709–715 (2004).
Weidinger, S. et al. Association study of mast cell chymase polymorphisms with atopy. Allergy 60, 1256–1261 (2005).
Sharma, S., Rajan, U.M., Kumar, A., Soni, A. & Ghosh, B. A novel (TG)n(GA)m repeat polymorphism 254 bp downstream of the mast cell chymase (CMA1) gene is associated with atopic asthma and total serum IgE levels. J. Hum. Genet. 50, 276–282 (2005).
Kim, S.H. et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin. Exp. Allergy 39, 203–212 (2009).
O'Regan, G.M., Sandilands, A., McLean, W.H. & Irvine, A.D. Filaggrin in atopic dermatitis. J. Allergy Clin. Immunol. 124, R2–R6 (2009).
Gan, S.Q., McBride, O.W., Idler, W.W., Markova, N. & Steinert, P.M. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry 29, 9432–9440 (1990).
Palmer, C.N. et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 38, 441–446 (2006).
Zhang, H. et al. Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy 66, 420–427 (2010).
Hanifin, J.M. et al. Diagnostic features of atopic dermatitis. Acta Dermatol. 92 Suppl, 44–47 (1980).
Krawczak, M. et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 9, 55–61 (2006).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Higgins, J.P.T. et al. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Mantel, N. et al. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 (1959).
Barrett, J.C. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Acknowledgements
We thank the individuals and their families who participated in this project. This study was funded by General Program of National Natural Science Foundation of China (81072461, 81071284, 30771196, 30771942, 30800990, 30800610) and the High-Tech Research and Development Program of China (863 Plan) (2007AA02Z161), the Key Project of Natural Science Foundation of China (30530670), Anhui Provincial Special Scientific Program (2007-7) and the National Basic Research Program of China (973 Plan) (2007B516801). S.W. is supported by a Heisenberg fellowship and a grant from the German Research Council DFG (WE2678/4–1, WE2678/6–1) as well as a grant from the German Ministry of Education and Research (BMBF) as part of the National Genome Research Network (NGFN grant 01GS 0818).
Author information
Authors and Affiliations
Contributions
X.-J.Z. conceived of this study and obtained financial support. X.-J.Z., Z.-R.Y., S.Y., L.-D.S., F.-L.X., Y. Li and W.-M.Z. participated in the design and were responsible for sample selection, genotyping and project management. H.Z., A.F., H.S., R.F.-H., S.W., M.S., Q.L., X.-L.C., Y.-F.G., Ming L., Y.-H.Z., J.-P.T., H.W., X.-Y.Z., X.-Y. Liu, L.M., A.-P.J., X.-H.G., C.-X.T., S.-P.S., Z.-Y.L., X.-L.Z., J.C., X.-Y. Luo, X.D., W.S., C.-P.S., X.-B.F., X.-P.H., J.-Q.H., Y.W., Y.-X.B., S.-M.H., Y.-M.L., K.-J.Z., D.-Y.H., Min L., X.Z., B.-R.G., Y. Liu, S.-M.Z., X.-Y.Y., Y.-Q.R., Y.C., M.G., P.-G.W., H.L., S.-X.L., C.-J.Y., G.-S.L., Z.-X.W., H.-Y.T. and X.F. conducted sample selection and data management, undertook recruitment, collected phenotype data, undertook related data handling and calculation, managed recruitment and obtained biological samples. F.-S.Z., G.C., X.-D.Z. and P.L. performed genotyping analysis. J.-J.L., H.-Y.T., X.-F.T. and X.-B.Z. undertook data processing, statistical analysis and bioinformatics investigations. All the authors contributed to the final paper, with X.-J.Z., Z.-R.Y., S.Y., L.-D.S., F.-L.X., Y. Li and W.-M.Z. having key roles.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–6. (PDF 875 kb)
Rights and permissions
About this article
Cite this article
Sun, LD., Xiao, FL., Li, Y. et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet 43, 690–694 (2011). https://doi.org/10.1038/ng.851
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.851
This article is cited by
-
European and multi-ancestry genome-wide association meta-analysis of atopic dermatitis highlights importance of systemic immune regulation
Nature Communications (2023)
-
Confirming the TMEM232 gene associated with atopic dermatitis through targeted capture sequencing
Scientific Reports (2021)
-
Association of Gasdermin B Gene GSDMB Polymorphisms with Risk of Allergic Diseases
Biochemical Genetics (2021)
-
Rare variant analysis in eczema identifies exonic variants in DUSP1, NOTCH4 and SLC9A4
Nature Communications (2021)
-
Methylome and transcriptome profiles in three yak tissues revealed that DNA methylation and the transcription factor ZGPAT co-regulate milk production
BMC Genomics (2020)