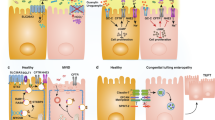
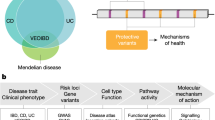
Abstract
Recent advances have provided substantial insight into the maintenance of mucosal immunity and the pathogenesis of inflammatory bowel disease. Cellular programs responsible for intestinal homeostasis use diverse intracellular and intercellular networks to promote immune tolerance, inflammation or epithelial restitution. Complex interfaces integrate local host and microbial signals to activate appropriate effector programs selectively and even drive plasticity between these programs. In addition, genetic studies and mouse models have emphasized the role of genetic predispositions and how they affect interactions with microbial and environmental factors, leading to pro-colitogenic perturbations of the host–commensal relationship.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature Genet. 42, 1118–1125 (2010). This study identifies 39 new loci containing genes in previously identified Crohn's-disease-relevant pathways, including IL10, IL27 and TYK2 , and reports the important genetic overlap with loci implicated in other immune-related diseases.
Anderson, C. A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature Genet. 43, 246–252 (2011). This study identifies 29 new loci containing genes that reinforce the contribution of epithelial barrier function, intracellular defence and cytokine-dependent signalling in ulcerative colitis, and it also reports genetic overlap with Crohn's disease.
Spehlmann, M. E. et al. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm. Bowel Dis. 14, 968–976 (2008).
Yazdanyar, S., Kamstrup, P. R., Tybjaerg-Hansen, A. & Nordestgaard, B. G. Penetrance of NOD2/CARD15 genetic variants in the general population. Can. Med. Assoc. J. 182, 661–665 (2010).
Monick, M. M. et al. Identification of an autophagy defect in smokers' alveolar macrophages. J. Immunol. 185, 5425–5435 (2010).
Momozawa, Y. et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nature Genet. 43, 43–47 (2010).
Wang, K. et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 19, 2059–2067 (2010).
Janse, M. et al. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL and CARD9. Hepatology 53, 1977–1985 (2011).
Zhang, F.-R. et al. Genomewide association study of leprosy. N. Engl. J. Med. 361, 2609–2618 (2009).
Waisberg, M. et al. Genetic susceptibility to systemic lupus erythematosus protects against cerebral malaria in mice. Proc. Natl Acad. Sci. USA 108, 1122–1127 (2011).
Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010).
Glocker, E.-O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Di Meglio, P. et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS ONE 6, e17160 (2011).
Yu, R. Y. & Gallagher, G. A naturally occurring, soluble antagonist of human IL-23 inhibits the development and in vitro function of human Th17 cells. J. Immunol. 185, 7302–7308 (2010).
Rosenstiel, P. et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc. Natl Acad. Sci. USA 103, 3280–3285 (2006).
Brest, P. et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nature Genet. 43, 242–245 (2011).
Khalil, A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009).
Muise, A. M. et al. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn's disease. Gut 58, 1121–1127 (2009).
Sabath, E. et al. Ga12 regulates protein interactions within the MDCK cell tight junction and inhibits tight-junction assembly. J. Cell Sci. 121, 814–824 (2008).
Scharl, M. et al. Protection of epithelial barrier function by the Crohn's disease associated gene protein tyrosine phosphatase N2. Gastroenterology 137, 2030–2040.e5 (2009).
Hassan, S. W. et al. Increased susceptibility to dextran sulfate sodium induced colitis in the T cell protein tyrosine phosphatase heterozygous mouse. PLoS ONE 5, e8868 (2010).
Darsigny, M. et al. Loss of hepatocyte-nuclear-factor-4α affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS ONE 4, e7609 (2009).
Cattin, A.-L. et al. Hepatocyte nuclear factor 4α, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol. 29, 6294–6308 (2009).
Pabst, O., Zweigerdt, R. & Arnold, H. H. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development 126, 2215–2225 (1999).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009).
Fu, J. et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Invest. 121, 1657–1666 (2011).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Biswas, A. et al. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc. Natl Acad. Sci. USA 107, 14739–14744 (2010).
Kaser, A. et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008). This study illustrates the role of Paneth and goblet cells in intestinal homeostasis and how the control of ER stress is crucial for their maintenance, such that XBP1 deficiency leads to Paneth cell loss, reduced goblet cell numbers and increased susceptibility to experimental colitis.
Brandl, K. et al. Enhanced sensitivity to DSS colitis caused by a hypomorphic Mbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proc. Natl Acad. Sci. USA 106, 3300–3305 (2009).
Goodall, J. C. et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc. Natl Acad. Sci. USA 107, 17698–17703 (2010).
Campbell, E. L. et al. Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc. Natl Acad. Sci. USA 107, 14298–14303 (2010).
Rescigno, M. Intestinal dendritic cells. Adv. Immunol. 107, 109–138 (2010).
Blasius, A. L. et al. Slc15a4, AP-3, and Hermansky–Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107, 19973–19978 (2010).
Smith, A. M. et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease. J. Exp. Med. 206, 1883–1897 (2009).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009). This pioneering study uses computational analyses of time-course gene-expression profiles to characterize dendritic cell programs and predict signalling circuits engaged after stimulation by microbe-associated molecules,and it exerimentally validates key regulators.
Siddiqui, K. R. R., Laffont, S. & Powrie, F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity 32, 557–567 (2010).
Uematsu, S. et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nature Immunol. 7, 868–874 (2006).
Swirski, F. K. et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616 (2009).
Sawa, S. et al. RORγ+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature Immunol. 12, 320–326 (2011).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009). This study describes mucosal IL-22-secreting ILCs in humans and mice, capturing many key properties of these cells, including their ability to respond to IL-23 stimulation and promote epithelial restitutive effects.
Takayama, T. et al. Imbalance of NKp44+NKp46− and NKp44−NKp46+ natural killer cells in the intestinal mucosa of patients with Crohn's disease. Gastroenterology 139, 882–892.e3 (2010).
Shaw, M. H., Kamada, N., Warner, N., Kim, Y. G. & Nunez, G. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol. 32, 73–79 (2011).
Cooney, R. et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Med. 16, 90–97 (2010).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 11, 55–62 (2010).
Sabbah, A. et al. Activation of innate immune antiviral responses by Nod2. Nature Immunol. 10, 1073–1080 (2009).
Zaki, M. H. et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010).
Hsu, Y.-M. S. et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nature Immunol. 8, 198–205 (2007).
Poeck, H. et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nature Immunol. 11, 63–69 (2010).
Gringhuis, S. I. et al. Selective C-Rel activation via Malt1 controls anti-fungal TH-17 immunity by dectin-1 and dectin-2. PLoS Pathogens 7, e1001259 (2011).
Schroeder, B. O. et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469, 419–423 (2011).
Dolowschiak, T. et al. Potentiation of epithelial innate host responses by intercellular communication. PLoS Pathogens 6, e1001194 (2010).
Gardet, A. et al. LRRK2 is involved in the IFN-γ response and host response to pathogens. J. Immunol. 185, 5577–5585 (2010).
Wu, W., Hsu, Y.-M. S., Bi, L., Songyang, Z. & Lin, X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nature Immunol. 10, 1208–1214 (2009).
Efimova, O., Szankasi, P. & Kelley, T. W. Ncf1 (p47phox) is essential for direct regulatory T cell mediated suppression of CD4+ effector T cells. PLoS ONE 6, e16013 (2011).
Kraaij, M. D. et al. Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc. Natl Acad. Sci. USA 107, 17686–17691 (2010).
Rioux, J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 39, 596–604 (2007).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 39, 207–211 (2007).
McCarroll, S. A. et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nature Genet. 40, 1107–1112 (2008).
Kuballa, P., Huett, A., Rioux, J. D., Daly, M. J. & Xavier, R. J. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE 3, e3391 (2008).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). This seminal paper reports that a Crohn's disease susceptibility gene requires interaction with environmental (microbial) cues to manifest a disease phenotype.
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008).
Cretney, E. et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature Immunol. 12, 304–311 (2011).
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010).
Do, J. S., Visperas, A., Dong, C., Baldwin, W. M. III & Min, B. Generation of colitogenic Th17 CD4 T cells is enhanced by IL-17+ γδ T cells. J. Immunol. 186, 4546–4550 (2011).
Park, S.-G. et al. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity 33, 791–803 (2010).
Chen, Y. et al. Foxp3+ regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity 34, 409–421 (2011).
Trifari, S., Kaplan, C. D., Tran, E. H., Crellin, N. K. & Spits, H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from TH-17, TH1 and TH2 cells. Nature Immunol. 10, 864–871 (2009).
Lohr, N. J. et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 86, 447–453 (2010).
Ramon, H. E. et al. The ubiquitin ligase adaptor Ndfip1 regulates T cell-mediated gastrointestinal inflammation and inflammatory bowel disease susceptibility. Mucosal Immunol. 4, 314–324 (2010).
Cox, J. H. et al. IL-27 promotes T cell-dependent colitis through multiple mechanisms. J. Exp. Med. 208, 115–123 (2011).
Ferreira, R. C. et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nature Genet. 42, 777–780 (2010).
Surolia, I. et al. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature 466, 243–247 (2010).
Blair, P. A. et al. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity 32, 129–140 (2010).
Konkel, J. E. et al. Control of the development of CD8αα+ intestinal intraepithelial lymphocytes by TGF-β. Nature Immunol. 12, 312–319 (2011).
Darmoise, A. et al. Lysosomal α-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity 33, 216–228 (2010).
Boasberg, P., Hamid, O. & O'Day, S. Ipilimumab: unleashing the power of the immune system through CTLA-4 blockade. Semin. Oncol. 37, 440–449 (2010).
Turnbaugh, P. J. et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl Acad. Sci. USA 107, 7503–7508 (2010).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). This paper shows how S . Typhimurium can use ROS generated during inflammation to form an electron acceptor that confers a growth advantage, illustrating how gut environment can affect microbial communities by providing a selective advantage to some bacterial species.
Wen, L. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Wei, M. et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nature Immunol. 12, 264–270 (2011).
Fuhrer, A. et al. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J. Exp. Med. 207, 2843–2854 (2010).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009). This paper highlights how commensal populations can modulate the host immune response, identifying a bacterial metabolic product (SCFA) and its receptor (GPR43), and shows that this pathway can ameliorate experimental colitis.
Mogi, C. et al. Involvement of proton-sensing TDAG8 in extracellular acidification-induced inhibition of proinflammatory cytokine production in peritoneal macrophages. J. Immunol. 182, 3243–3251 (2009).
Fujiya, M. et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1, 299–308 (2007).
Sasawatari, S. et al. The solute carrier family 15A4 regulates TLR9 and NOD1 functions in the innate immune system and promotes colitis in mice. Gastroenterology 140, 1513–1525 (2011).
Brandl, K. et al. MyD88 signaling in nonhematopoietic cells protects mice against induced colitis by regulating specific EGF receptor ligands. Proc. Natl Acad. USA 107, 19967–19972 (2010).
Kobayashi, T. et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J. Immunol. 186, 4649–4655 (2011).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). This important study shows how particular components of the gut microbiota can modulate the adaptive immune response, with SFB promoting differentiation of pro-inflammatory T H 17 cells.
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2010). This study demonstrates that Clostridium clusters IV and XIVa affect the adaptive immune system by inducing a TGF-β-rich milieu and promoting differentiation of anti-inflammatory T reg cells.
Grainger, J. R. et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-β pathway. J. Exp. Med. 207, 2331–2341 (2010).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Acknowledgements
R.J.X., A.G. and B.K. are supported by grants from the National Institutes of Health and the Crohn's and Colitis Foundation of America. We apologize to those whose work is not cited owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Supplementary information
Supplementary References
This file contains a Supplementary Bibliography to the main paper. (PDF 231 kb)
Rights and permissions
About this article
Cite this article
Khor, B., Gardet, A. & Xavier, R. Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). https://doi.org/10.1038/nature10209
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10209
This article is cited by
-
Galectin from Trichinella spiralis alleviates DSS-induced colitis in mice by regulating the intestinal microbiota
Veterinary Research (2024)
-
An atypical presentation of ulcerative colitis: case report
Egyptian Pediatric Association Gazette (2024)
-
Miniaturized implantable temperature sensors for the long-term monitoring of chronic intestinal inflammation
Nature Biomedical Engineering (2024)
-
Discovering biomarkers associated and predicting cardiovascular disease with high accuracy using a novel nexus of machine learning techniques for precision medicine
Scientific Reports (2024)
-
Sesame cake extract attenuates dextran sulfate sodium-induced colitis through inhibition of oxidative stress in mice
Food Science and Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.